Lecture 6. Neural Networks#
How to train your neurons
Joaquin Vanschoren
Show code cell source
# Auto-setup when running on Google Colab
import os
if 'google.colab' in str(get_ipython()) and not os.path.exists('/content/master'):
!git clone -q https://github.com/ML-course/master.git /content/master
!pip --quiet install -r /content/master/requirements_colab.txt
%cd master/notebooks
# Global imports and settings
%matplotlib inline
from preamble import *
interactive = False # Set to True for interactive plots
if interactive:
fig_scale = 0.5
plt.rcParams.update(print_config)
else: # For printing
fig_scale = 0.4
plt.rcParams.update(print_config)
#HTML('''<style>.reveal pre code {max-height: 1000px !important;}</style>''')
Overview#
Neural architectures
Training neural nets
Forward pass: Tensor operations
Backward pass: Backpropagation
Neural network design:
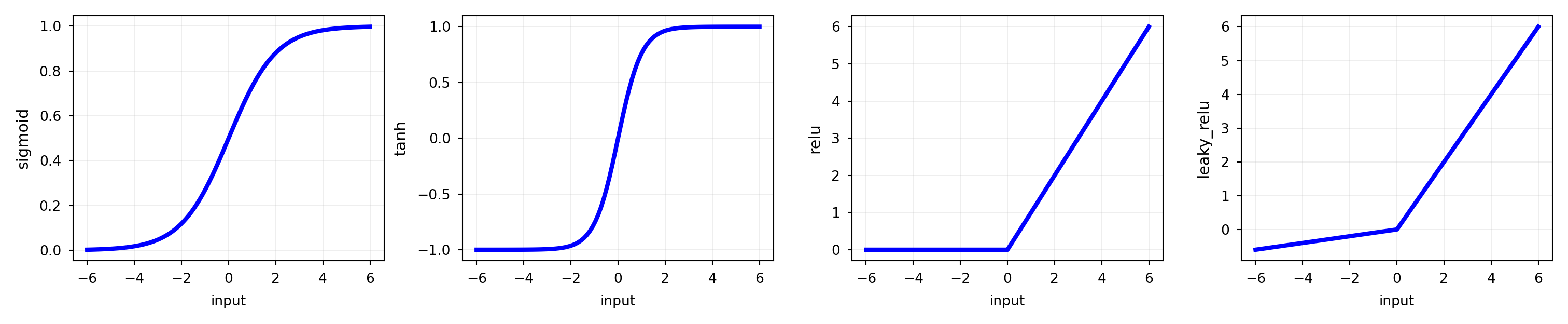
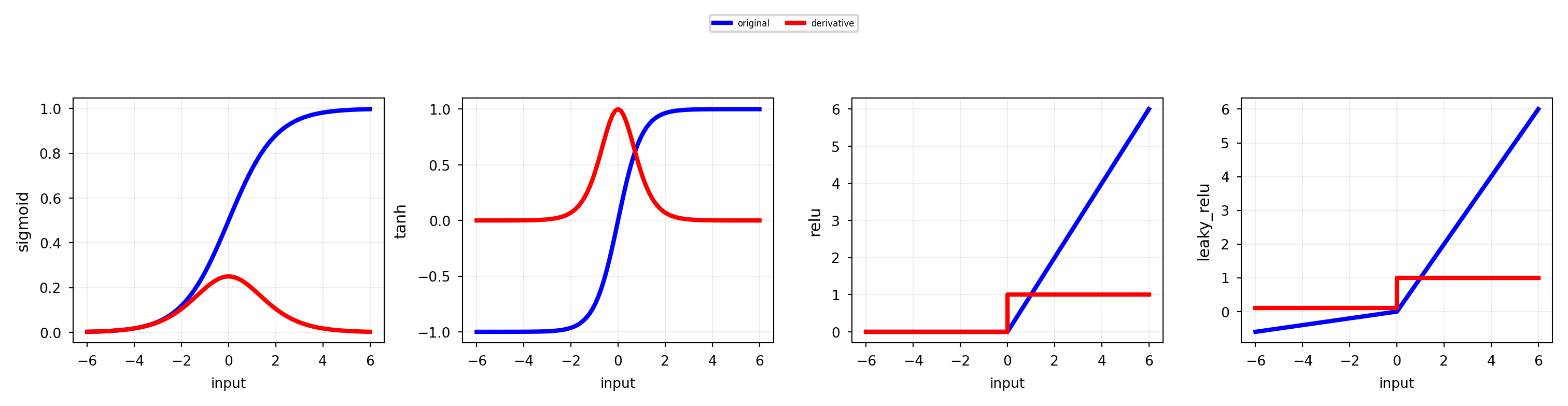
Activation functions
Weight initialization
Optimizers
Neural networks in practice
Model selection
Early stopping
Memorization capacity and information bottleneck
L1/L2 regularization
Dropout
Batch normalization
Show code cell source
def draw_neural_net(ax, layer_sizes, draw_bias=False, labels=False, activation=False, sigmoid=False,
weight_count=False, random_weights=False, show_activations=False, figsize=(4, 4)):
"""
Draws a dense neural net for educational purposes
Parameters:
ax: plot axis
layer_sizes: array with the sizes of every layer
draw_bias: whether to draw bias nodes
labels: whether to draw labels for the weights and nodes
activation: whether to show the activation function inside the nodes
sigmoid: whether the last activation function is a sigmoid
weight_count: whether to show the number of weights and biases
random_weights: whether to show random weights as colored lines
show_activations: whether to show a variable for the node activations
scale_ratio: ratio of the plot dimensions, e.g. 3/4
"""
figsize = (figsize[0]*fig_scale, figsize[1]*fig_scale)
left, right, bottom, top = 0.1, 0.89*figsize[0]/figsize[1], 0.1, 0.89
n_layers = len(layer_sizes)
v_spacing = (top - bottom)/float(max(layer_sizes))
h_spacing = (right - left)/float(len(layer_sizes) - 1)
colors = ['greenyellow','cornflowerblue','lightcoral']
w_count, b_count = 0, 0
ax.set_xlim(0, figsize[0]/figsize[1])
ax.axis('off')
ax.set_aspect('equal')
txtargs = {"fontsize":12*fig_scale, "verticalalignment":'center', "horizontalalignment":'center', "zorder":5}
# Draw biases by adding a node to every layer except the last one
if draw_bias:
layer_sizes = [x+1 for x in layer_sizes]
layer_sizes[-1] = layer_sizes[-1] - 1
# Nodes
for n, layer_size in enumerate(layer_sizes):
layer_top = v_spacing*(layer_size - 1)/2. + (top + bottom)/2.
node_size = v_spacing/len(layer_sizes) if activation and n!=0 else v_spacing/3.
if n==0:
color = colors[0]
elif n==len(layer_sizes)-1:
color = colors[2]
else:
color = colors[1]
for m in range(layer_size):
ax.add_artist(plt.Circle((n*h_spacing + left, layer_top - m*v_spacing), radius=node_size,
color=color, ec='k', zorder=4, linewidth=fig_scale))
b_count += 1
nx, ny = n*h_spacing + left, layer_top - m*v_spacing
nsx, nsy = [n*h_spacing + left,n*h_spacing + left], [layer_top - m*v_spacing - 0.5*node_size*2,layer_top - m*v_spacing + 0.5*node_size*2]
if draw_bias and m==0 and n<len(layer_sizes)-1:
ax.text(nx, ny, r'$1$', **txtargs)
elif labels and n==0:
ax.text(n*h_spacing + left,layer_top + v_spacing/1.5, 'input', **txtargs)
ax.text(nx, ny, r'$x_{}$'.format(m), **txtargs)
elif labels and n==len(layer_sizes)-1:
if activation:
if sigmoid:
ax.text(n*h_spacing + left,layer_top - m*v_spacing, r"$z \;\;\; \sigma$", **txtargs)
else:
ax.text(n*h_spacing + left,layer_top - m*v_spacing, r"$z_{} \;\; g$".format(m), **txtargs)
ax.add_artist(plt.Line2D(nsx, nsy, c='k', zorder=6))
if show_activations:
ax.text(n*h_spacing + left + 1.5*node_size,layer_top - m*v_spacing, r"$\hat{y}$", fontsize=12*fig_scale,
verticalalignment='center', horizontalalignment='left', zorder=5, c='r')
else:
ax.text(nx, ny, r'$o_{}$'.format(m), **txtargs)
ax.text(n*h_spacing + left,layer_top + v_spacing, 'output', **txtargs)
elif labels:
if activation:
ax.text(n*h_spacing + left,layer_top - m*v_spacing, r"$z_{} \;\; f$".format(m), **txtargs)
ax.add_artist(plt.Line2D(nsx, nsy, c='k', zorder=6))
if show_activations:
ax.text(n*h_spacing + left + node_size*1.2 ,layer_top - m*v_spacing, r"$a_{}$".format(m), fontsize=12*fig_scale,
verticalalignment='center', horizontalalignment='left', zorder=5, c='b')
else:
ax.text(nx, ny, r'$h_{}$'.format(m), **txtargs)
# Edges
for n, (layer_size_a, layer_size_b) in enumerate(zip(layer_sizes[:-1], layer_sizes[1:])):
layer_top_a = v_spacing*(layer_size_a - 1)/2. + (top + bottom)/2.
layer_top_b = v_spacing*(layer_size_b - 1)/2. + (top + bottom)/2.
for m in range(layer_size_a):
for o in range(layer_size_b):
if not (draw_bias and o==0 and len(layer_sizes)>2 and n<layer_size_b-1):
xs = [n*h_spacing + left, (n + 1)*h_spacing + left]
ys = [layer_top_a - m*v_spacing, layer_top_b - o*v_spacing]
color = 'k' if not random_weights else plt.cm.bwr(np.random.random())
ax.add_artist(plt.Line2D(xs, ys, c=color, alpha=0.6))
if not (draw_bias and m==0):
w_count += 1
if labels and not random_weights:
wl = r'$w_{{{},{}}}$'.format(m,o) if layer_size_b>1 else r'$w_{}$'.format(m)
ax.text(xs[0]+np.diff(xs)/2, np.mean(ys)-np.diff(ys)/9, wl, ha='center', va='center',
fontsize=12*fig_scale)
# Count
if weight_count:
b_count = b_count - layer_sizes[0]
if draw_bias:
b_count = b_count - (len(layer_sizes) - 2)
ax.text(right*1.05, bottom, "{} weights, {} biases".format(w_count, b_count), ha='center', va='center')
Architecture#
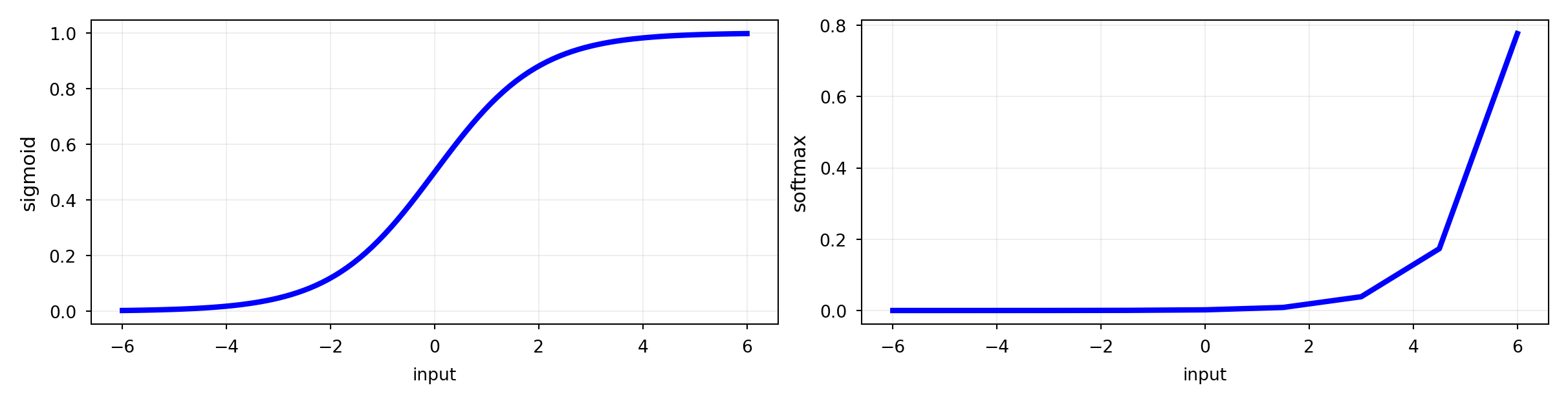
Logistic regression, drawn in a different, neuro-inspired, way
Linear model: inner product (\(z\)) of input vector \(\mathbf{x}\) and weight vector \(\mathbf{w}\), plus bias \(w_0\)
Logistic (or sigmoid) function maps the output to a probability in [0,1]
Uses log loss (cross-entropy) and gradient descent to learn the weights
Show code cell source
fig = plt.figure(figsize=(3*fig_scale,3*fig_scale))
ax = fig.gca()
draw_neural_net(ax, [4, 1], activation=True, draw_bias=True, labels=True, sigmoid=True)
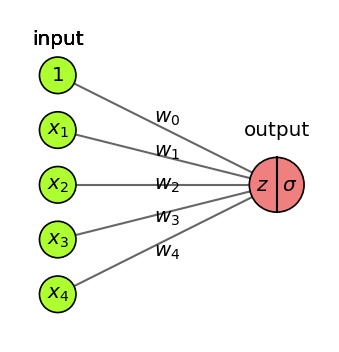
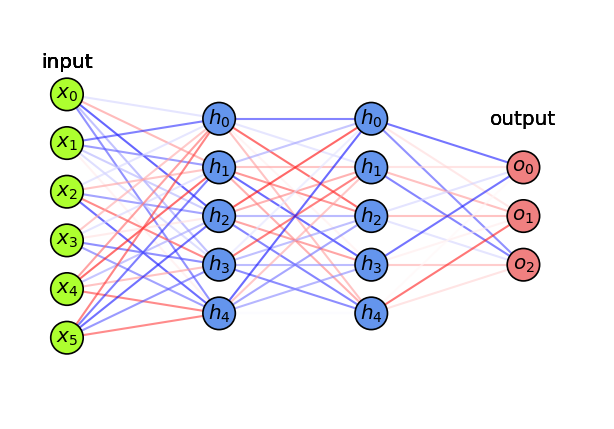
Basic Architecture#
Add one (or more) hidden layers \(h\) with \(k\) nodes (or units, cells, neurons)
Every ‘neuron’ is a tiny function, the network is an arbitrarily complex function
Weights \(w_{i,j}\) between node \(i\) and node \(j\) form a weight matrix \(\mathbf{W}^{(l)}\) per layer \(l\)
Every neuron weights the inputs \(\mathbf{x}\) and passes it through a non-linear activation function
Activation functions (\(f,g\)) can be different per layer, output \(\mathbf{a}\) is called activation $\(\color{blue}{h(\mathbf{x})} = \color{blue}{\mathbf{a}} = f(\mathbf{z}) = f(\mathbf{W}^{(1)} \color{green}{\mathbf{x}}+\mathbf{w}^{(1)}_0) \quad \quad \color{red}{o(\mathbf{x})} = g(\mathbf{W}^{(2)} \color{blue}{\mathbf{a}}+\mathbf{w}^{(2)}_0)\)$
Show code cell source
fig, axes = plt.subplots(1,2, figsize=(10*fig_scale,5*fig_scale))
draw_neural_net(axes[0], [2, 3, 1], draw_bias=True, labels=True, weight_count=True, figsize=(4, 4))
draw_neural_net(axes[1], [2, 3, 1], activation=True, show_activations=True, draw_bias=True,
labels=True, weight_count=True, figsize=(4, 4))
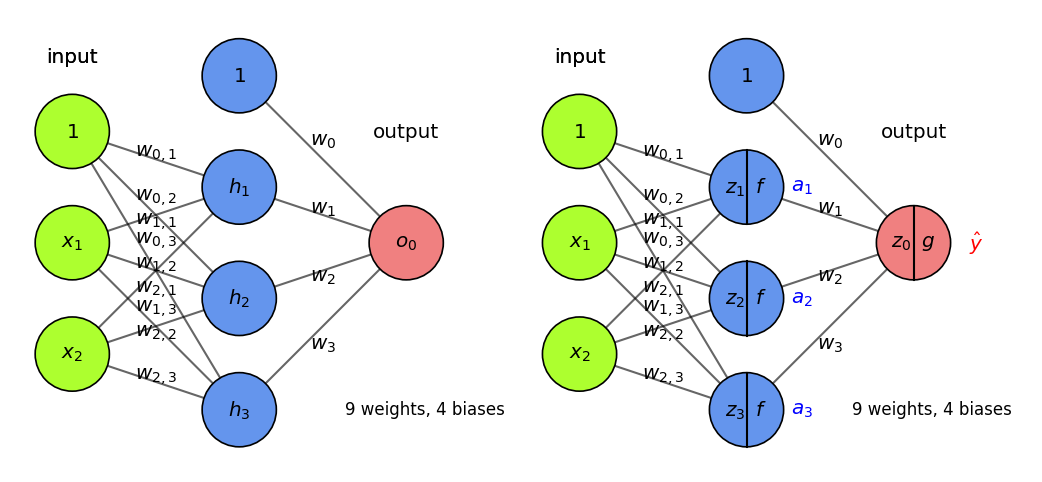
More layers#
Add more layers, and more nodes per layer, to make the model more complex
For simplicity, we don’t draw the biases (but remember that they are there)
In dense (fully-connected) layers, every previous layer node is connected to all nodes
The output layer can also have multiple nodes (e.g. 1 per class in multi-class classification)
Show code cell source
import ipywidgets as widgets
from ipywidgets import interact, interact_manual
@interact
def plot_dense_net(nr_layers=(0,6,1), nr_nodes=(1,12,1)):
fig = plt.figure(figsize=(10*fig_scale, 5*fig_scale))
ax = fig.gca()
ax.axis('off')
hidden = [nr_nodes]*nr_layers
draw_neural_net(ax, [5] + hidden + [5], weight_count=True, figsize=(6, 4))
plt.show()
Show code cell source
if not interactive:
plot_dense_net(nr_layers=6, nr_nodes=10)
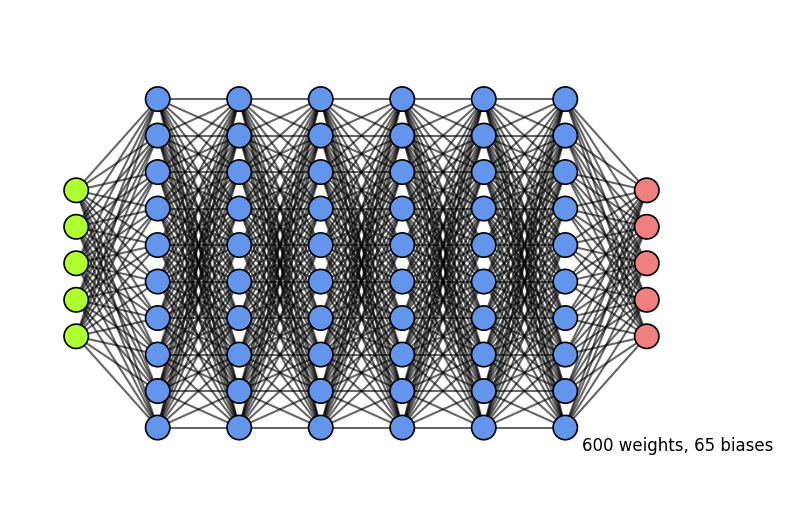
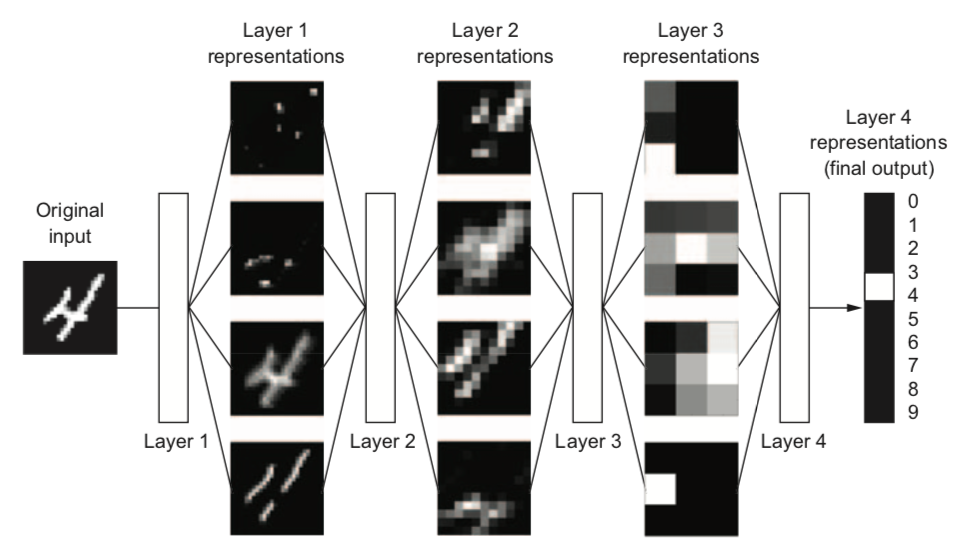
Why layers?#
Each layer acts as a filter and learns a new representation of the data
Subsequent layers can learn iterative refinements
Easier than learning a complex relationship in one go
Example: for image input, each layer yields new (filtered) images
Can learn multiple mappings at once: weight tensor \(\mathit{W}\) yields activation tensor \(\mathit{A}\)
From low-level patterns (edges, end-points, …) to combinations thereof
Each neuron ‘lights up’ if certain patterns occur in the input
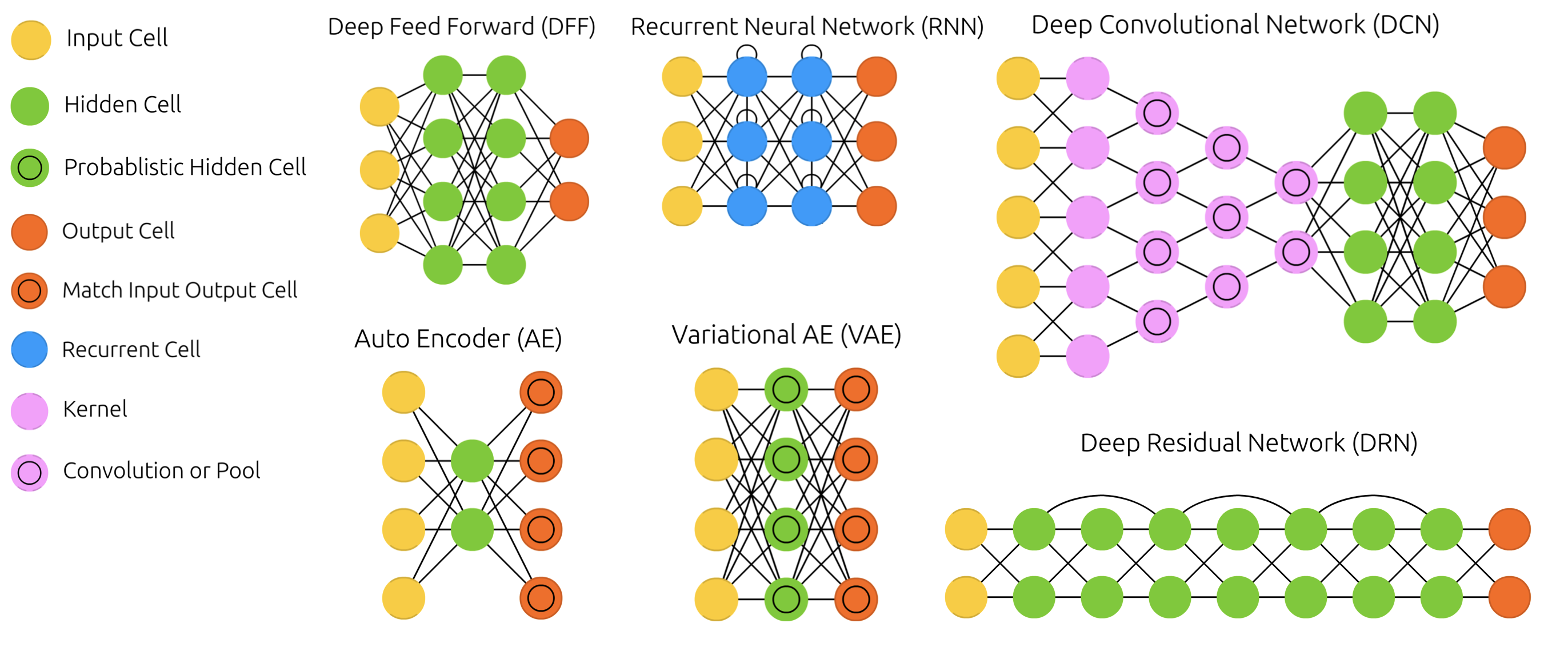
Other architectures#
There exist MANY types of networks for many different tasks
Convolutional nets for image data, Recurrent nets for sequential data,…
Also used to learn representations (embeddings), generate new images, text,…
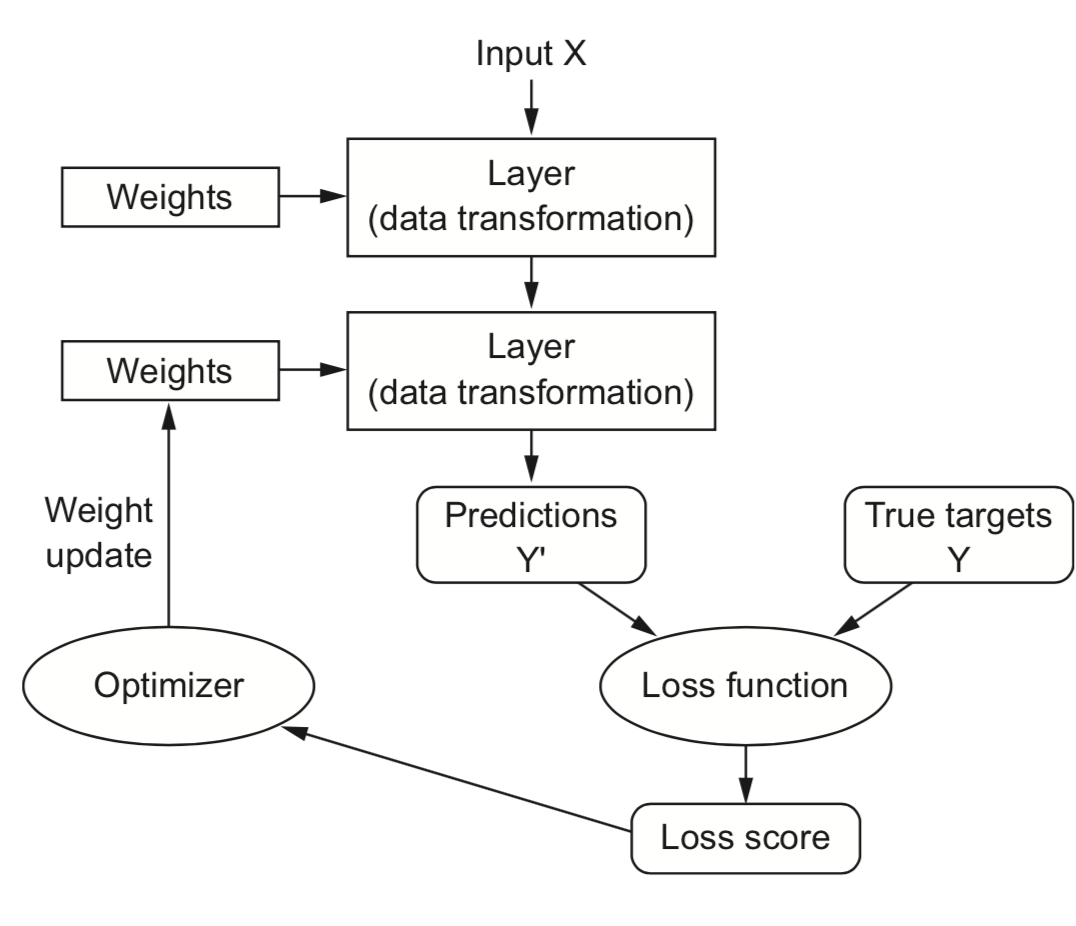
Training Neural Nets#
Design the architecture, choose activation functions (e.g. sigmoids)
Choose a way to initialize the weights (e.g. random initialization)
Choose a loss function (e.g. log loss) to measure how well the model fits training data
Choose an optimizer (typically an SGD variant) to update the weights
Mini-batch Stochastic Gradient Descent (recap)#
Draw a batch of batch_size training data \(\mathbf{X}\) and \(\mathbf{y}\)
Forward pass : pass \(\mathbf{X}\) though the network to yield predictions \(\mathbf{\hat{y}}\)
Compute the loss \(\mathcal{L}\) (mismatch between \(\mathbf{\hat{y}}\) and \(\mathbf{y}\))
Backward pass : Compute the gradient of the loss with regard to every weight
Backpropagate the gradients through all the layers
Update \(W\): \(W_{(i+1)} = W_{(i)} - \frac{\partial L(x, W_{(i)})}{\partial W} * \eta\)
Repeat until n passes (epochs) are made through the entire training set
Show code cell source
# TODO: show the actual weight updates
@interact
def draw_updates(iteration=(1,100,1)):
fig, ax = plt.subplots(1, 1, figsize=(6*fig_scale, 4*fig_scale))
np.random.seed(iteration)
draw_neural_net(ax, [6,5,5,3], labels=True, random_weights=True, show_activations=True, figsize=(6, 4));
plt.show()
Show code cell source
if not interactive:
draw_updates(iteration=1)
Forward pass#
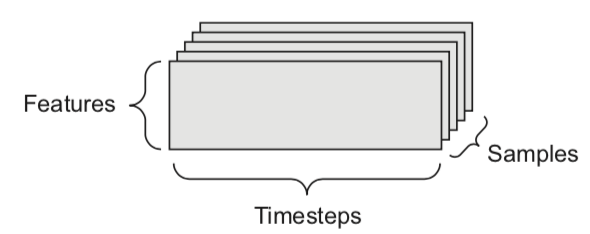
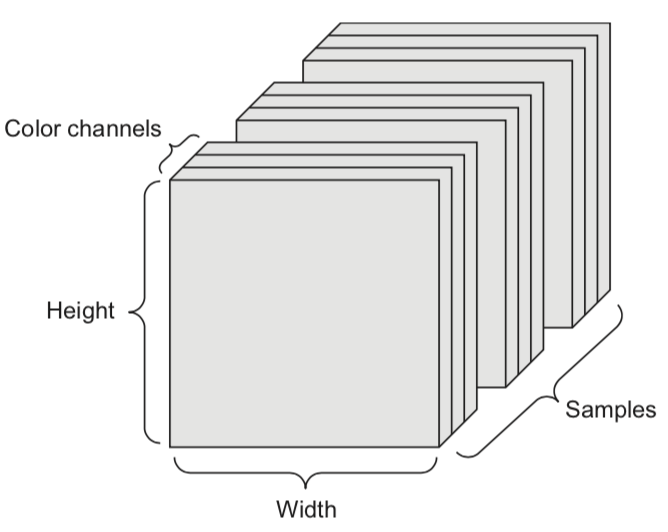
We can naturally represent the data as tensors
Numerical n-dimensional array (with n axes)
2D tensor: matrix (samples, features)
3D tensor: time series (samples, timesteps, features)
4D tensor: color images (samples, height, width, channels)
5D tensor: video (samples, frames, height, width, channels)
Tensor operations#
The operations that the network performs on the data can be reduced to a series of tensor operations
These are also much easier to run on GPUs
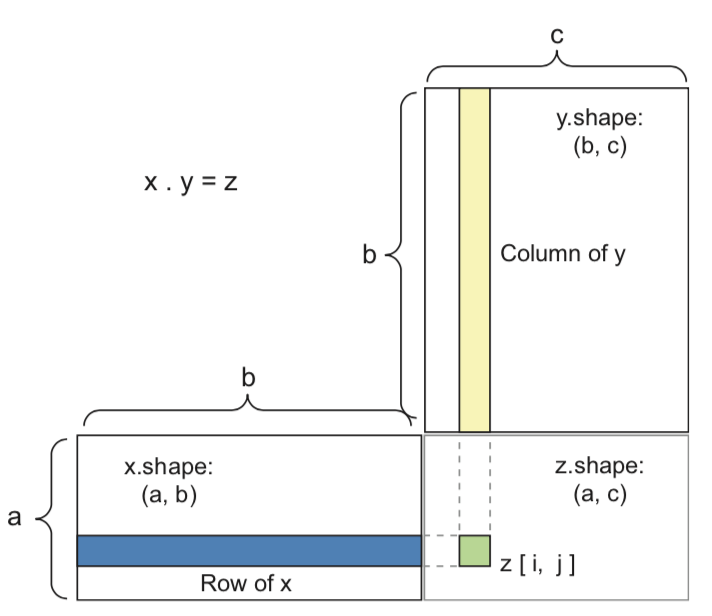
A dense layer with sigmoid activation, input tensor \(\mathbf{X}\), weight tensor \(\mathbf{W}\), bias \(\mathbf{b}\):
y = sigmoid(np.dot(X, W) + b)
Tensor dot product for 2D inputs (\(a\) samples, \(b\) features, \(c\) hidden nodes)
Element-wise operations#
Activation functions and addition are element-wise operations:
def sigmoid(x):
return 1/(1 + np.exp(-x))
def add(x, y):
return x + y
Note: if y has a lower dimension than x, it will be broadcasted: axes are added to match the dimensionality, and y is repeated along the new axes
>>> np.array([[1,2],[3,4]]) + np.array([10,20])
array([[11, 22],
[13, 24]])
Backward pass (backpropagation)#
For last layer, compute gradient of the loss function \(\mathcal{L}\) w.r.t all weights of layer \(l\)
Sum up the gradients for all \(\mathbf{x}_j\) in minibatch: \(\sum_{j} \frac{\partial \mathcal{L}(\mathbf{x}_j,y_j)}{\partial W^{(l)}}\)
Update all weights in a layer at once (with learning rate \(\eta\)): \(W_{(i+1)}^{(l)} = W_{(i)}^{(l)} - \eta \sum_{j} \frac{\partial \mathcal{L}(\mathbf{x}_j,y_j)}{\partial W_{(i)}^{(l)}}\)
Repeat for next layer, iterating backwards (most efficient, avoids redundant calculations)
Example#
Imagine feeding a single data point, output is \(\hat{y} = g(z) = g(w_0 + w_1 * a_1 + w_2 * a_2 +... + w_p * a_p)\)
Decrease loss by updating weights:
Update the weights of last layer to maximize improvement: \(w_{i,(new)} = w_{i} - \frac{\partial \mathcal{L}}{\partial w_i} * \eta\)
To compute gradient \(\frac{\partial \mathcal{L}}{\partial w_i}\) we need the chain rule: \(f(g(x)) = f'(g(x)) * g'(x)\) $\(\frac{\partial \mathcal{L}}{\partial w_i} = \color{red}{\frac{\partial \mathcal{L}}{\partial g}} \color{blue}{\frac{\partial \mathcal{g}}{\partial z_0}} \color{green}{\frac{\partial \mathcal{z_0}}{\partial w_i}}\)$
E.g., with \(\mathcal{L} = \frac{1}{2}(y-\hat{y})^2\) and sigmoid \(\sigma\): \(\frac{\partial \mathcal{L}}{\partial w_i} = \color{red}{(y - \hat{y})} * \color{blue}{\sigma'(z_0)} * \color{green}{a_i}\)
Show code cell source
fig = plt.figure(figsize=(4*fig_scale, 3.5*fig_scale))
ax = fig.gca()
draw_neural_net(ax, [2, 3, 1], activation=True, draw_bias=True, labels=True,
show_activations=True)
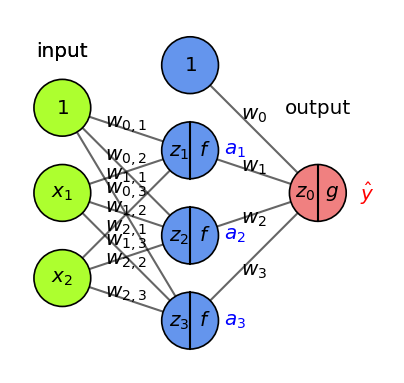
Backpropagation (2)#
Another way to decrease the loss \(\mathcal{L}\) is to update the activations \(a_i\)
To update \(a_i = f(z_i)\), we need to update the weights of the previous layer
We want to nudge \(a_i\) in the right direction by updating \(w_{i,j}\): $\(\frac{\partial \mathcal{L}}{\partial w_{i,j}} = \frac{\partial \mathcal{L}}{\partial a_i} \frac{\partial a_i}{\partial z_i} \frac{\partial \mathcal{z_i}}{\partial w_{i,j}} = \left( \frac{\partial \mathcal{L}}{\partial g} \frac{\partial \mathcal{g}}{\partial z_0} \frac{\partial \mathcal{z_0}}{\partial a_i} \right) \frac{\partial a_i}{\partial z_i} \frac{\partial \mathcal{z_i}}{\partial w_{i,j}}\)$
We know \(\frac{\partial \mathcal{L}}{\partial g}\) and \(\frac{\partial \mathcal{g}}{\partial z_0}\) from the previous step, \(\frac{\partial \mathcal{z_0}}{\partial a_i} = w_i\), \(\frac{\partial a_i}{\partial z_i} = f'\) and \(\frac{\partial \mathcal{z_i}}{\partial w_{i,j}} = x_j\)
Show code cell source
fig = plt.figure(figsize=(4*fig_scale, 4*fig_scale))
ax = fig.gca()
draw_neural_net(ax, [2, 3, 1], activation=True, draw_bias=True, labels=True,
show_activations=True)
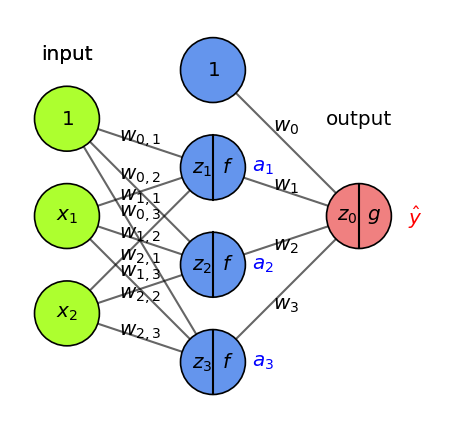
Backpropagation (3)#
With multiple output nodes, \(\mathcal{L}\) is the sum of all per-output (per-class) losses
\(\frac{\partial \mathcal{L}}{\partial a_i}\) is sum of the gradients for every output
Per layer, sum up gradients for every point \(\mathbf{x}\) in the batch: \(\sum_{j} \frac{\partial \mathcal{L}(\mathbf{x}_j,y_j)}{\partial W}\)
Update all weights of every layer \(l\)
\(W_{(i+1)}^{(l)} = W_{(i)}^{(l)} - \eta \sum_{j} \frac{\partial \mathcal{L}(\mathbf{x}_j,y_j)}{\partial W_{(i)}^{(l)}}\)
Repeat with a new batch of data until loss converges
Show code cell source
fig = plt.figure(figsize=(8*fig_scale, 4*fig_scale))
ax = fig.gca()
draw_neural_net(ax, [2, 3, 3, 2], activation=True, draw_bias=True, labels=True,
random_weights=True, show_activations=True, figsize=(8, 4))
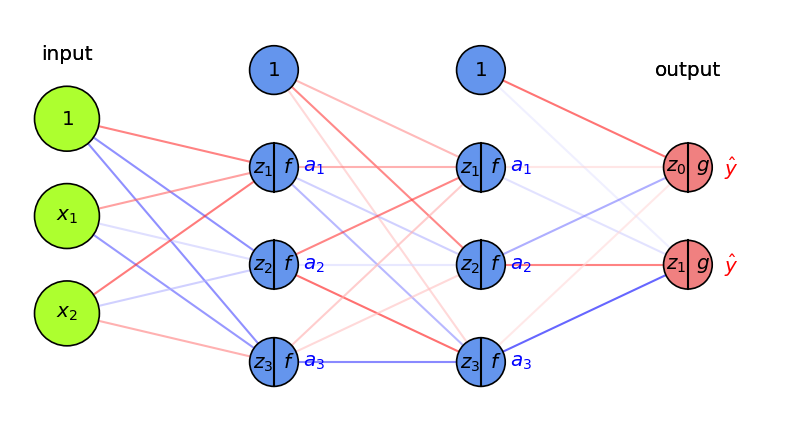
Summary#
The network output \(a_o\) is defined by the weights \(W^{(o)}\) and biases \(\mathbf{b}^{(o)}\) of the output layer, and
The activations of a hidden layer \(h_1\) with activation function \(a_{h_1}\), weights \(W^{(1)}\) and biases \(\mathbf{b^{(1)}}\):
Minimize the loss by SGD. For layer \(l\), compute \(\frac{\partial \mathcal{L}(a_o(x))}{\partial W_l}\) and \(\frac{\partial \mathcal{L}(a_o(x))}{\partial b_{l,i}}\) using the chain rule
Decomposes into gradient of layer above, gradient of activation function, gradient of layer input:
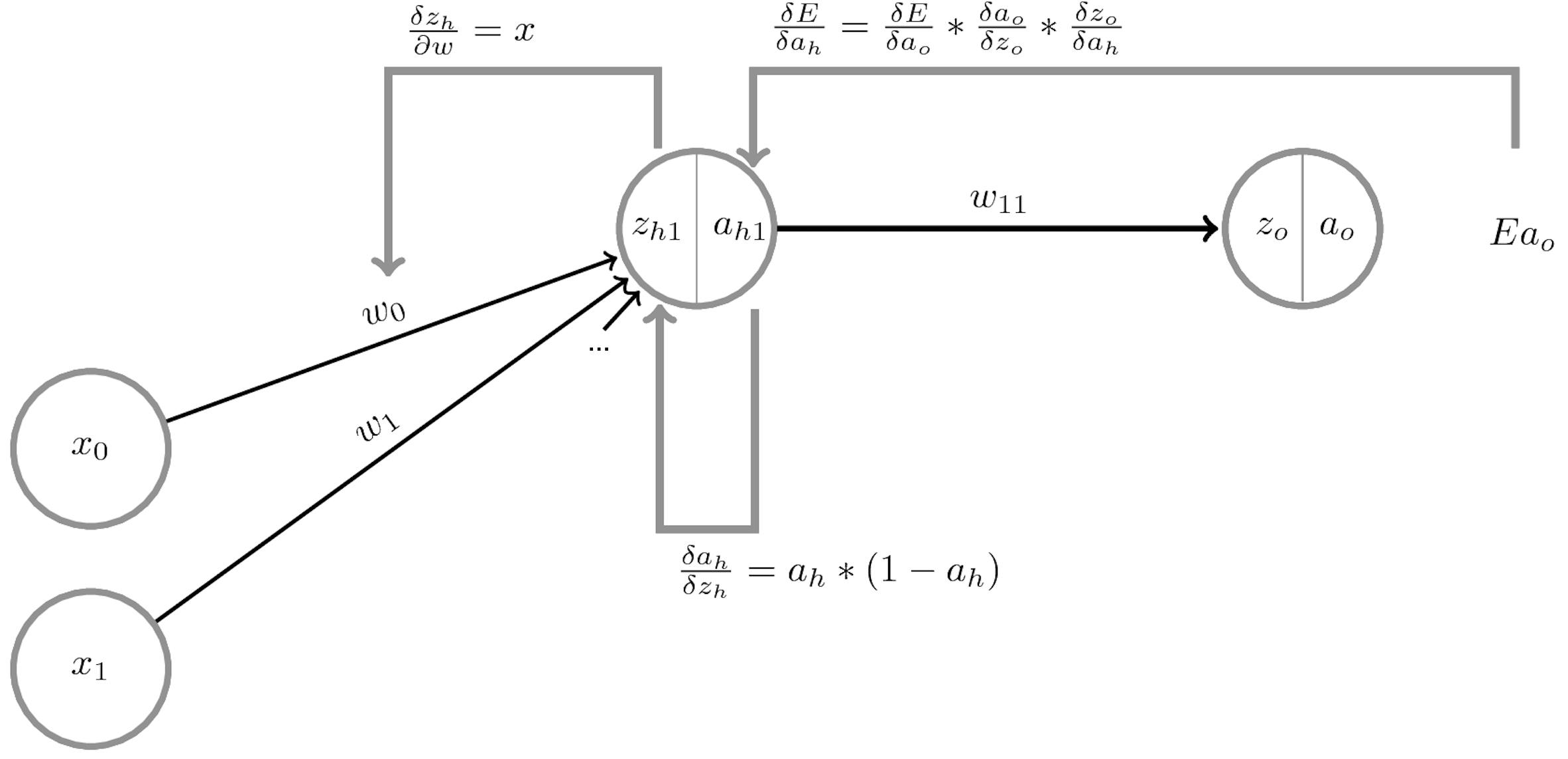
Weight initialization#
Initializing weights to 0 is bad: all gradients in layer will be identical (symmetry)
Too small random weights shrink activations to 0 along the layers (vanishing gradient)
Too large random weights multiply along layers (exploding gradient, zig-zagging)
Ideal: small random weights + variance of input and output gradients remains the same
Glorot/Xavier initialization (for tanh): randomly sample from \(N(0,\sigma), \sigma = \sqrt{\frac{2}{\text{fan_in + fan_out}}}\)
fan_in: number of input units, fan_out: number of output units
He initialization (for ReLU): randomly sample from \(N(0,\sigma), \sigma = \sqrt{\frac{2}{\text{fan_in}}}\)
Uniform sampling (instead of \(N(0,\sigma)\)) for deeper networks (w.r.t. vanishing gradients)
Show code cell source
fig, ax = plt.subplots(1,1, figsize=(6*fig_scale, 3*fig_scale))
draw_neural_net(ax, [3, 5, 5, 5, 5, 5, 3], random_weights=True, figsize=(6, 3))
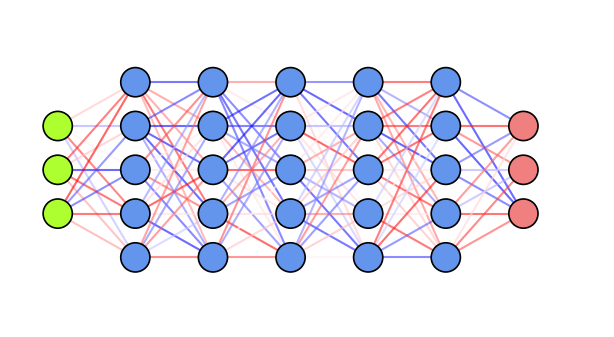
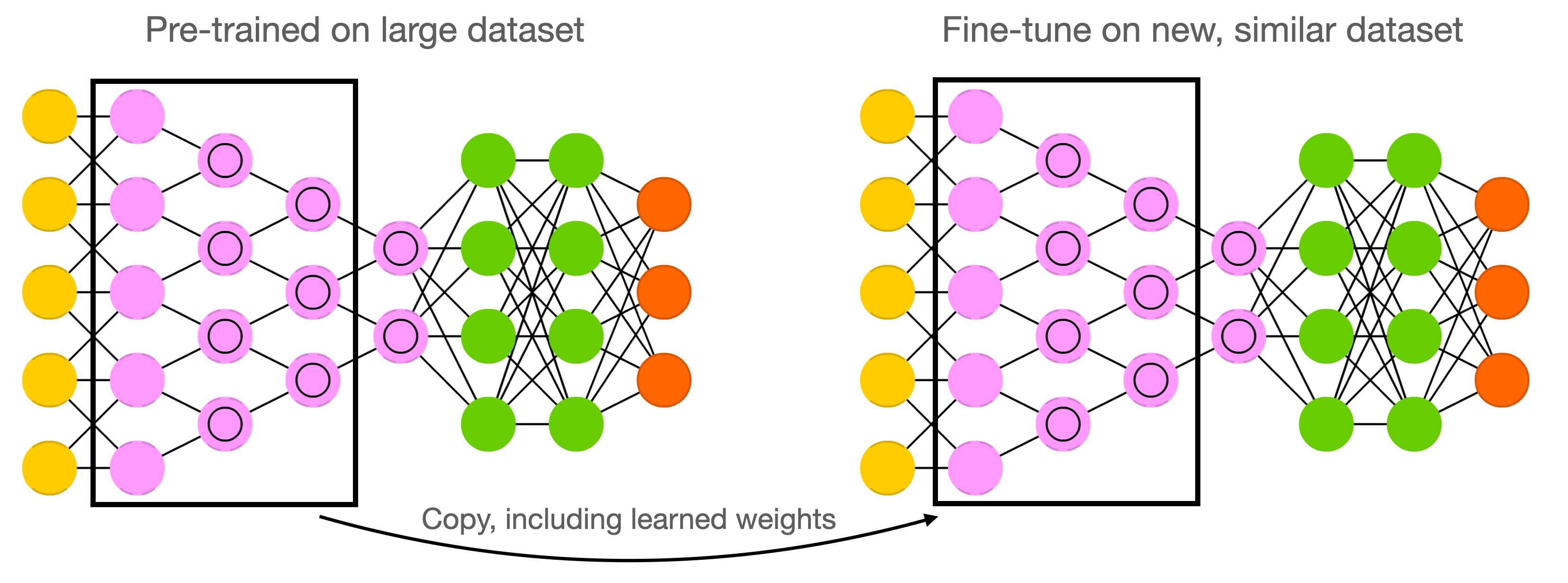
Weight initialization: transfer learning#
Instead of starting from scratch, start from weights previously learned from similar tasks
This is, to a big extent, how humans learn so fast
Transfer learning: learn weights on task T, transfer them to new network
Weights can be frozen, or finetuned to the new data
Only works if the previous task is ‘similar’ enough
Generally, weights learned on very diverse data (e.g. ImageNet) transfer better
Meta-learning: learn a good initialization across many related tasks
import tensorflow as tf
#import tensorflow_addons as tfa
# Toy surface
def f(x, y):
return (1.5 - x + x*y)**2 + (2.25 - x + x*y**2)**2 + (2.625 - x + x*y**3)**2
# Tensorflow optimizers
class CyclicalLearningRate(tf.keras.optimizers.schedules.LearningRateSchedule):
def __init__(self, initial_learning_rate, maximal_learning_rate, step_size):
self.initial_learning_rate = initial_learning_rate
self.maximal_learning_rate = maximal_learning_rate
self.step_size = step_size
def __call__(self, step):
cycle = tf.floor(1 + step / (2 * self.step_size))
x = tf.abs(step / self.step_size - 2 * cycle + 1)
return self.initial_learning_rate + (self.maximal_learning_rate - self.initial_learning_rate) * tf.maximum(0.0, (1 - x))
clr_schedule = CyclicalLearningRate(initial_learning_rate=1e-4,
maximal_learning_rate=0.1,
step_size=100)
sgd_cyclic = tf.keras.optimizers.SGD(learning_rate=clr_schedule)
sgd = tf.optimizers.SGD(0.01)
lr_schedule = tf.optimizers.schedules.ExponentialDecay(0.02,decay_steps=100,decay_rate=0.96)
sgd_decay = tf.optimizers.SGD(learning_rate=lr_schedule)
momentum = tf.optimizers.SGD(0.005, momentum=0.9, nesterov=False)
nesterov = tf.optimizers.SGD(0.005, momentum=0.9, nesterov=True)
adagrad = tf.optimizers.Adagrad(0.4)
rmsprop = tf.optimizers.RMSprop(learning_rate=0.1)
adam = tf.optimizers.Adam(learning_rate=0.2, beta_1=0.9, beta_2=0.999, epsilon=1e-8)
optimizers = [sgd, sgd_decay, momentum, nesterov, adagrad, rmsprop, adam, sgd_cyclic]
opt_names = ['sgd', 'sgd_decay', 'momentum', 'nesterov', 'adagrad', 'rmsprop', 'adam', 'sgd_cyclic']
cmap = plt.cm.get_cmap('tab10')
colors = [cmap(x/10) for x in range(10)]
# Training
all_paths = []
for opt, name in zip(optimizers, opt_names):
x = tf.Variable(0.8)
y = tf.Variable(1.6)
x_history = []
y_history = []
loss_prev = 0.0
max_steps = 100
for step in range(max_steps):
with tf.GradientTape() as g:
loss = f(x, y)
x_history.append(x.numpy())
y_history.append(y.numpy())
grads = g.gradient(loss, [x, y])
opt.apply_gradients(zip(grads, [x, y]))
if np.abs(loss_prev - loss.numpy()) < 1e-6:
break
loss_prev = loss.numpy()
x_history = np.array(x_history)
y_history = np.array(y_history)
path = np.concatenate((np.expand_dims(x_history, 1), np.expand_dims(y_history, 1)), axis=1).T
all_paths.append(path)
# Plotting
number_of_points = 50
margin = 4.5
minima = np.array([3., .5])
minima_ = minima.reshape(-1, 1)
x_min = 0. - 2
x_max = 0. + 3.5
y_min = 0. - 3.5
y_max = 0. + 2
x_points = np.linspace(x_min, x_max, number_of_points)
y_points = np.linspace(y_min, y_max, number_of_points)
x_mesh, y_mesh = np.meshgrid(x_points, y_points)
z = np.array([f(xps, yps) for xps, yps in zip(x_mesh, y_mesh)])
def plot_optimizers(ax, iterations, optimizers):
ax.contour(x_mesh, y_mesh, z, levels=np.logspace(-0.5, 5, 25), norm=LogNorm(), cmap=plt.cm.jet, linewidths=fig_scale, zorder=-1)
ax.plot(*minima, 'r*', markersize=20*fig_scale)
for name, path, color in zip(opt_names, all_paths, colors):
if name in optimizers:
p = path[:,:iterations]
ax.plot([], [], color=color, label=name, lw=3*fig_scale, linestyle='-')
ax.quiver(p[0,:-1], p[1,:-1], p[0,1:]-p[0,:-1], p[1,1:]-p[1,:-1], scale_units='xy', angles='xy', scale=1, color=color, lw=4)
ax.set_xlim((x_min, x_max))
ax.set_ylim((y_min, y_max))
ax.legend(loc='lower left', prop={'size': 15*fig_scale})
ax.set_xticks([])
ax.set_yticks([])
plt.tight_layout()
from decimal import *
from matplotlib.colors import LogNorm
# Training for momentum
all_lr_paths = []
lr_range = [0.005 * i for i in range(0,10)]
for lr in lr_range:
opt = tf.optimizers.SGD(lr, nesterov=False)
x_init = 0.8
x = tf.compat.v1.get_variable('x', dtype=tf.float32, initializer=tf.constant(x_init))
y_init = 1.6
y = tf.compat.v1.get_variable('y', dtype=tf.float32, initializer=tf.constant(y_init))
x_history = []
y_history = []
z_prev = 0.0
max_steps = 100
for step in range(max_steps):
with tf.GradientTape() as g:
z = f(x, y)
x_history.append(x.numpy())
y_history.append(y.numpy())
dz_dx, dz_dy = g.gradient(z, [x, y])
opt.apply_gradients(zip([dz_dx, dz_dy], [x, y]))
if np.abs(z_prev - z.numpy()) < 1e-6:
break
z_prev = z.numpy()
x_history = np.array(x_history)
y_history = np.array(y_history)
path = np.concatenate((np.expand_dims(x_history, 1), np.expand_dims(y_history, 1)), axis=1).T
all_lr_paths.append(path)
# Plotting
number_of_points = 50
margin = 4.5
minima = np.array([3., .5])
minima_ = minima.reshape(-1, 1)
x_min = 0. - 2
x_max = 0. + 3.5
y_min = 0. - 3.5
y_max = 0. + 2
x_points = np.linspace(x_min, x_max, number_of_points)
y_points = np.linspace(y_min, y_max, number_of_points)
x_mesh, y_mesh = np.meshgrid(x_points, y_points)
z = np.array([f(xps, yps) for xps, yps in zip(x_mesh, y_mesh)])
def plot_learning_rate_optimizers(ax, iterations, lr):
ax.contour(x_mesh, y_mesh, z, levels=np.logspace(-0.5, 5, 25), norm=LogNorm(), cmap=plt.cm.jet, linewidths=fig_scale, zorder=-1)
ax.plot(*minima, 'r*', markersize=20*fig_scale)
for path, lrate in zip(all_lr_paths, lr_range):
if round(lrate,3) == lr:
p = path[:,:iterations]
ax.plot([], [], color='b', label="Learning rate {}".format(lr), lw=3*fig_scale, linestyle='-')
ax.quiver(p[0,:-1], p[1,:-1], p[0,1:]-p[0,:-1], p[1,1:]-p[1,:-1], scale_units='xy', angles='xy', scale=1, color='b', lw=4)
ax.set_xlim((x_min, x_max))
ax.set_ylim((y_min, y_max))
ax.legend(loc='lower left', prop={'size': 15*fig_scale})
ax.set_xticks([])
ax.set_yticks([])
plt.tight_layout()
Show code cell source
# Toy plot to illustrate nesterov momentum
# TODO: replace with actual gradient computation?
def plot_nesterov(ax, method="Nesterov momentum"):
ax.contour(x_mesh, y_mesh, z, levels=np.logspace(-0.5, 5, 25), norm=LogNorm(), cmap=plt.cm.jet, linewidths=fig_scale, zorder=-1)
ax.plot(*minima, 'r*', markersize=20*fig_scale)
# toy example
ax.quiver(-0.8,-1.13,1,1.33, scale_units='xy', angles='xy', scale=1, color='k', alpha=0.5, lw=3, label="previous update")
# 0.9 * previous update
ax.quiver(0.2,0.2,0.9,1.2, scale_units='xy', angles='xy', scale=1, color='g', lw=3, label="momentum step")
if method == "Momentum":
ax.quiver(0.2,0.2,0.5,0, scale_units='xy', angles='xy', scale=1, color='r', lw=3, label="gradient step")
ax.quiver(0.2,0.2,0.9*0.9+0.5,1.2, scale_units='xy', angles='xy', scale=1, color='b', lw=3, label="actual step")
if method == "Nesterov momentum":
ax.quiver(1.1,1.4,-0.2,-1, scale_units='xy', angles='xy', scale=1, color='r', lw=3, label="'lookahead' gradient step")
ax.quiver(0.2,0.2,0.7,0.2, scale_units='xy', angles='xy', scale=1, color='b', lw=3, label="actual step")
ax.set_title(method)
ax.set_xlim((x_min, x_max))
ax.set_ylim((-2.5, y_max))
ax.legend(loc='lower right', prop={'size': 9*fig_scale})
ax.set_xticks([])
ax.set_yticks([])
plt.tight_layout()
Optimizers#
SGD with learning rate schedules#
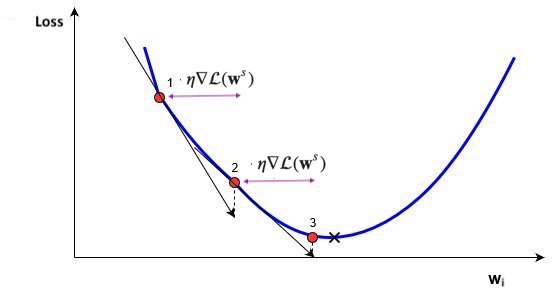
Using a constant learning \(\eta\) rate for weight updates \(\mathbf{w}_{(s+1)} = \mathbf{w}_s-\eta\nabla \mathcal{L}(\mathbf{w}_s)\) is not ideal
You would need to ‘magically’ know the right value
import ipywidgets as widgets
from ipywidgets import interact, interact_manual
from IPython.display import clear_output
@interact
def plot_lr(iterations=(1,100,1), learning_rate=(0.005,0.04,0.005)):
fig, ax = plt.subplots(figsize=(6*fig_scale,4*fig_scale))
plot_learning_rate_optimizers(ax,iterations,round(learning_rate,3))
plt.show()
if not interactive:
plot_lr(iterations=50, learning_rate=0.02)
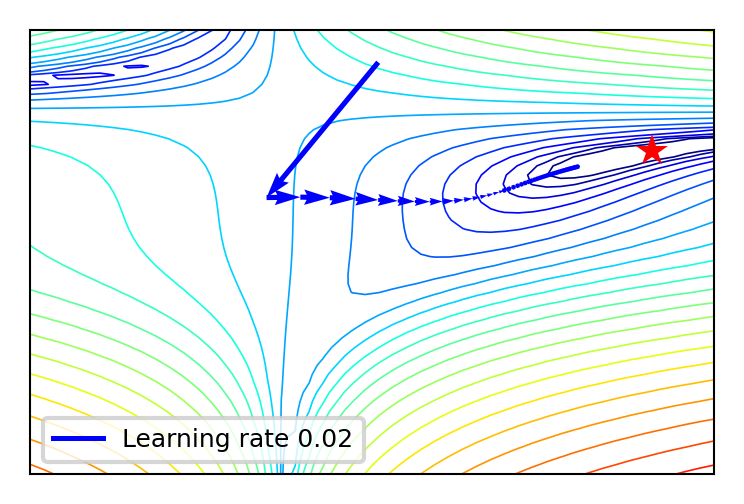
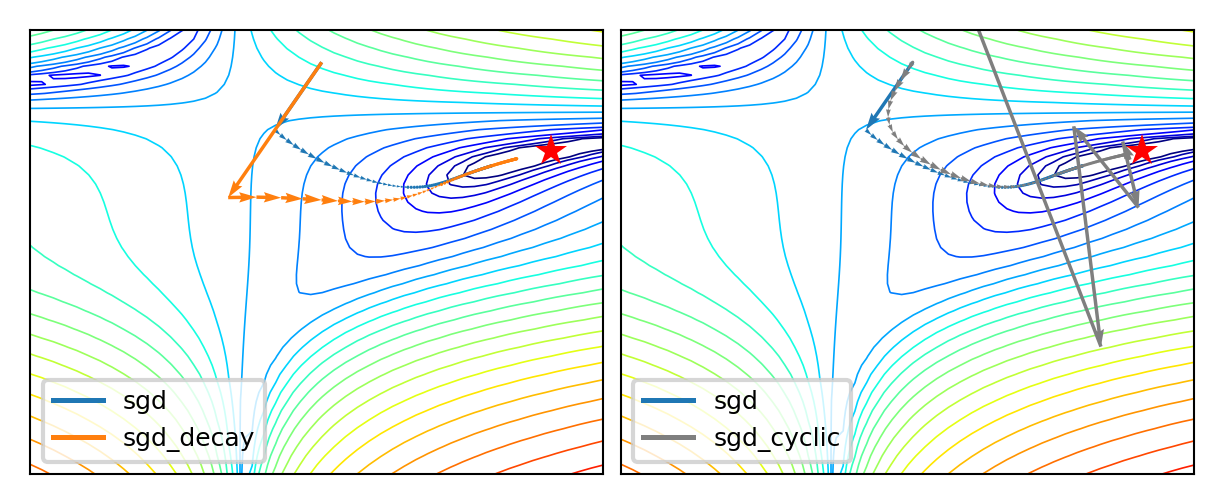
SGD with learning rate schedules#
Learning rate decay/annealing with decay rate \(k\)
E.g. exponential (\(\eta_{s+1} = \eta_{0} e^{-ks}\)), inverse-time (\(\eta_{s+1} = \frac{\eta_{0}}{1+ks}\)),…
Cyclical learning rates
Change from small to large: hopefully in ‘good’ region long enough before diverging
Warm restarts: aggressive decay + reset to initial learning rate
Show code cell source
@interact
def compare_optimizers(iterations=(1,100,1), optimizer1=opt_names, optimizer2=opt_names):
fig, ax = plt.subplots(figsize=(6*fig_scale,4*fig_scale))
plot_optimizers(ax,iterations,[optimizer1,optimizer2])
plt.show()
Show code cell source
if not interactive:
fig, axes = plt.subplots(1,2, figsize=(10*fig_scale,4*fig_scale))
optimizers = ['sgd_decay', 'sgd_cyclic']
for function, ax in zip(optimizers,axes):
plot_optimizers(ax,100,function)
plt.tight_layout();
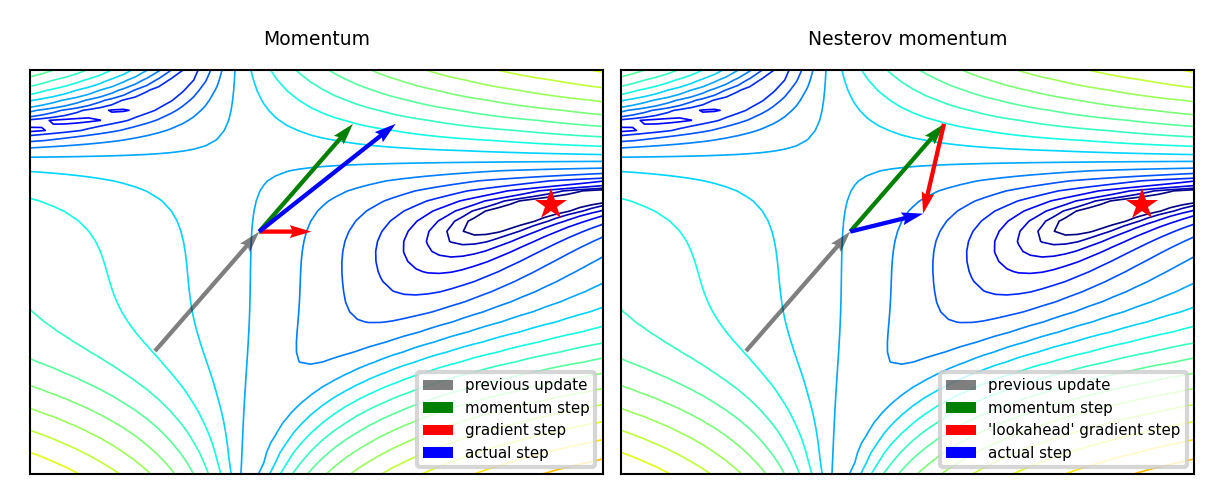
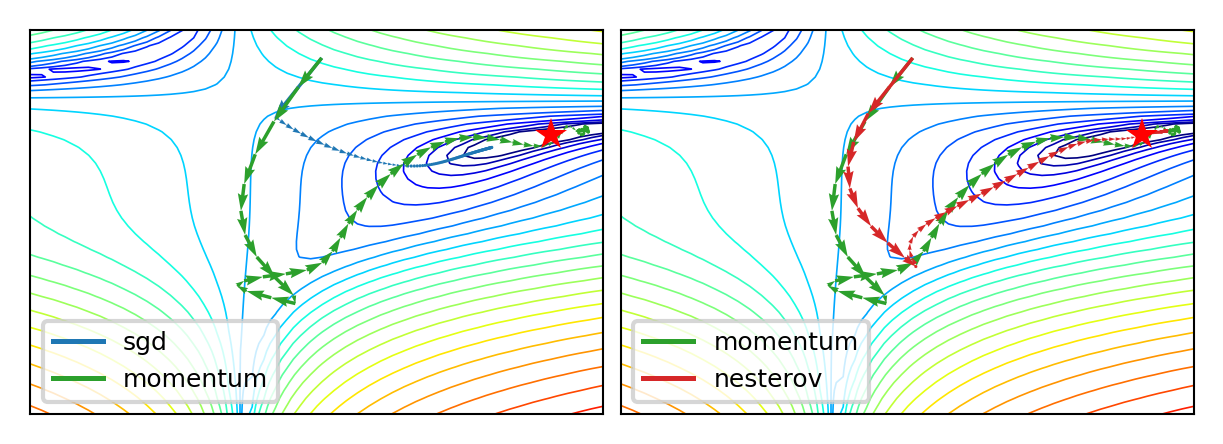
Momentum#
Imagine a ball rolling downhill: accumulates momentum, doesn’t exactly follow steepest descent
Reduces oscillation, follows larger (consistent) gradient of the loss surface
Adds a velocity vector \(\mathbf{v}\) with momentum \(\gamma\) (e.g. 0.9, or increase from \(\gamma=0.5\) to \(\gamma=0.99\)) $\(\mathbf{w}_{(s+1)} = \mathbf{w}_{(s)} + \mathbf{v}_{(s)} \qquad \text{with} \qquad \color{blue}{\mathbf{v}_{(s)}} = \color{green}{\gamma \mathbf{v}_{(s-1)}} - \color{red}{\eta \nabla \mathcal{L}(\mathbf{w}_{(s)})}\)$
Nesterov momentum: Look where momentum step would bring you, compute gradient there
Responds faster (and reduces momentum) when the gradient changes $\(\color{blue}{\mathbf{v}_{(s)}} = \color{green}{\gamma \mathbf{v}_{(s-1)}} - \color{red}{\eta \nabla \mathcal{L}(\mathbf{w}_{(s)} + \gamma \mathbf{v}_{(s-1)})}\)$
Show code cell source
fig, axes = plt.subplots(1,2, figsize=(10*fig_scale,4*fig_scale))
plot_nesterov(axes[0],method="Momentum")
plot_nesterov(axes[1],method="Nesterov momentum")
Momentum in practice#
Show code cell source
@interact
def compare_optimizers(iterations=(1,100,1), optimizer1=opt_names, optimizer2=opt_names):
fig, ax = plt.subplots(figsize=(6*fig_scale,4*fig_scale))
plot_optimizers(ax,iterations,[optimizer1,optimizer2])
plt.show()
Show code cell source
if not interactive:
fig, axes = plt.subplots(1,2, figsize=(10*fig_scale,3.5*fig_scale))
optimizers = [['sgd','momentum'], ['momentum','nesterov']]
for function, ax in zip(optimizers,axes):
plot_optimizers(ax,100,function)
plt.tight_layout();
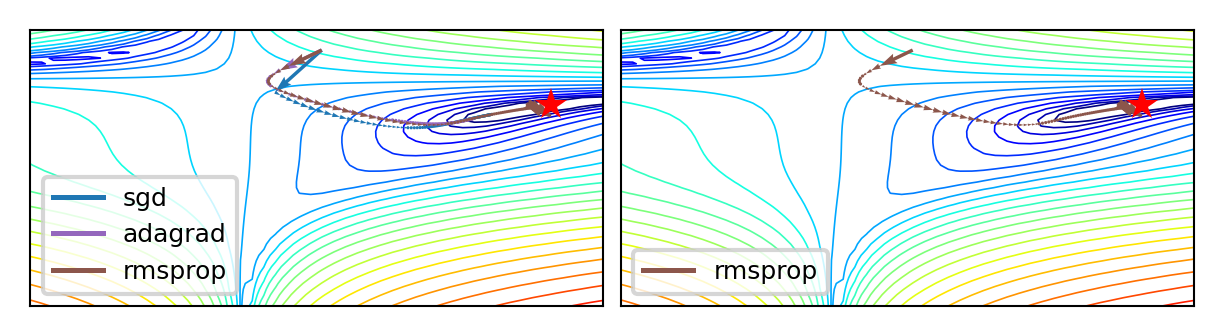
Adaptive gradients#
‘Correct’ the learning rate for each \(w_i\) based on specific local conditions (layer depth, fan-in,…)
Adagrad: scale \(\eta\) according to squared sum of previous gradients \(G_{i,(s)} = \sum_{t=1}^s \nabla \mathcal{L}(w_{i,(t)})^2\)
Update rule for \(w_i\). Usually \(\epsilon=10^{-7}\) (avoids division by 0), \(\eta=0.001\). $\(w_{i,(s+1)} = w_{i,(s)} - \frac{\eta}{\sqrt{G_{i,(s)}+\epsilon}} \nabla \mathcal{L}(w_{i,(s)})\)$
RMSProp: use moving average of squared gradients \(m_{i,(s)} = \gamma m_{i,(s-1)} + (1-\gamma) \nabla \mathcal{L}(w_{i,(s)})^2\)
Avoids that gradients dwindle to 0 as \(G_{i,(s)}\) grows. Usually \(\gamma=0.9, \eta=0.001\) $\(w_{i,(s+1)} = w_{i,(s)}- \frac{\eta}{\sqrt{m_{i,(s)}+\epsilon}} \nabla \mathcal{L}(w_{i,(s)})\)$
Show code cell source
if not interactive:
fig, axes = plt.subplots(1,2, figsize=(10*fig_scale,2.6*fig_scale))
optimizers = [['sgd','adagrad', 'rmsprop'], ['rmsprop','rmsprop_mom']]
for function, ax in zip(optimizers,axes):
plot_optimizers(ax,100,function)
plt.tight_layout();
Show code cell source
@interact
def compare_optimizers(iterations=(1,100,1), optimizer1=opt_names, optimizer2=opt_names):
fig, ax = plt.subplots(figsize=(6*fig_scale,4*fig_scale))
plot_optimizers(ax,iterations,[optimizer1,optimizer2])
plt.show()
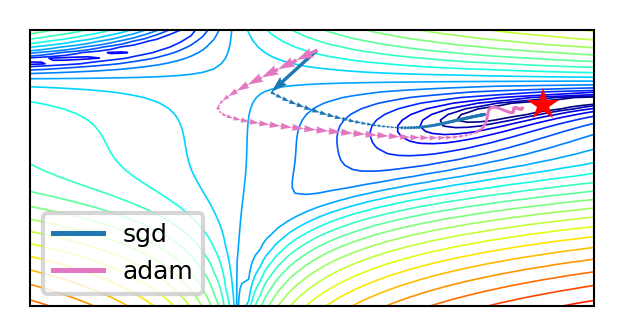
Adam (Adaptive moment estimation)#
Adam: RMSProp + momentum. Adds moving average for gradients as well (\(\gamma_2\) = momentum):
Adds a bias correction to avoid small initial gradients: \(\hat{m}_{i,(s)} = \frac{m_{i,(s)}}{1-\gamma}\) and \(\hat{g}_{i,(s)} = \frac{g_{i,(s)}}{1-\gamma_2}\) $\(g_{i,(s)} = \gamma_2 g_{i,(s-1)} + (1-\gamma_2) \nabla \mathcal{L}(w_{i,(s)})\)\( \)\(w_{i,(s+1)} = w_{i,(s)}- \frac{\eta}{\sqrt{\hat{m}_{i,(s)}+\epsilon}} \hat{g}_{i,(s)}\)$
Adamax: Idem, but use max() instead of moving average: \(u_{i,(s)} = max(\gamma u_{i,(s-1)}, |\mathcal{L}(w_{i,(s)})|)\) $\(w_{i,(s+1)} = w_{i,(s)}- \frac{\eta}{u_{i,(s)}} \hat{g}_{i,(s)}\)$
Show code cell source
if not interactive:
# fig, axes = plt.subplots(1,2, figsize=(10*fig_scale,2.6*fig_scale))
# optimizers = [['sgd','adam'], ['adam','adamax']]
# for function, ax in zip(optimizers,axes):
# plot_optimizers(ax,100,function)
# plt.tight_layout();
fig, axes = plt.subplots(1,1, figsize=(5*fig_scale,2.6*fig_scale))
optimizers = [['sgd','adam']]
plot_optimizers(axes,100,['sgd','adam'])
plt.tight_layout();
Show code cell source
@interact
def compare_optimizers(iterations=(1,100,1), optimizer1=opt_names, optimizer2=opt_names):
fig, ax = plt.subplots(figsize=(6*fig_scale,4*fig_scale))
plot_optimizers(ax,iterations,[optimizer1,optimizer2])
plt.show()
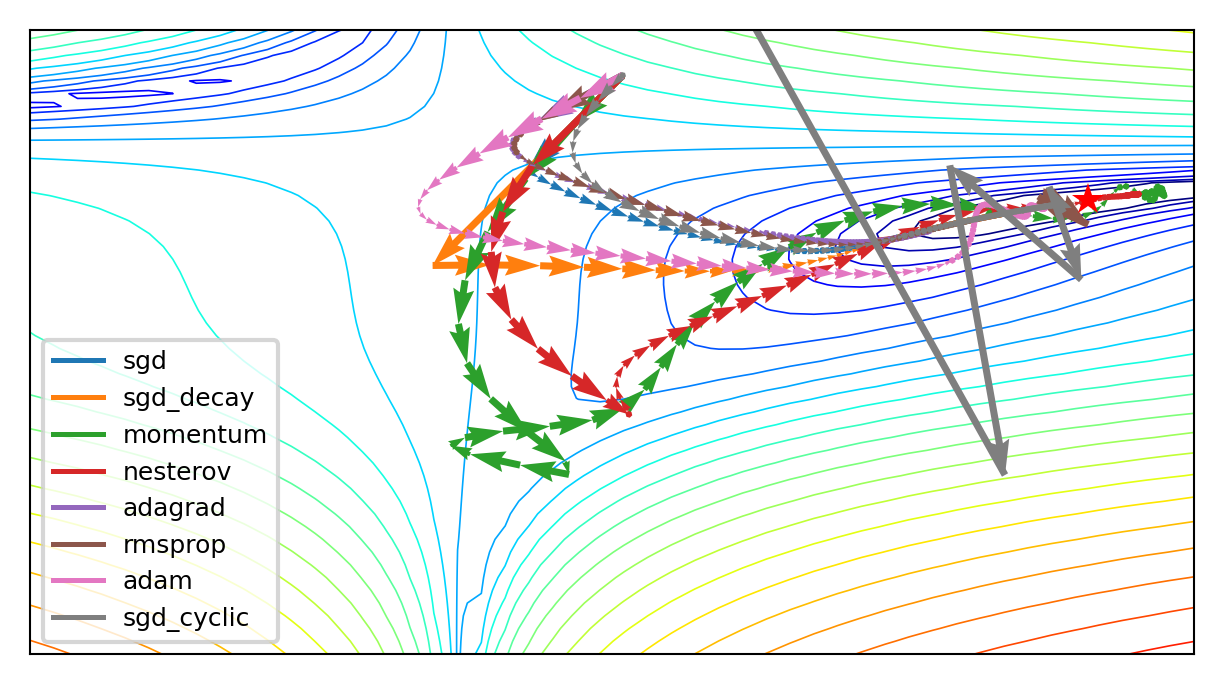
SGD Optimizer Zoo#
RMSProp often works well, but do try alternatives. For even more optimizers, see here.
Show code cell source
if not interactive:
fig, ax = plt.subplots(1,1, figsize=(10*fig_scale,5.5*fig_scale))
plot_optimizers(ax,100,opt_names)
Show code cell source
@interact
def compare_optimizers(iterations=(1,100,1)):
fig, ax = plt.subplots(figsize=(10*fig_scale,6*fig_scale))
plot_optimizers(ax,iterations,opt_names)
plt.show()
Show code cell source
from tensorflow.keras import models
from tensorflow.keras import layers
from numpy.random import seed
from tensorflow.random import set_seed
import random
import os
#Trying to set all seeds
os.environ['PYTHONHASHSEED']=str(0)
random.seed(0)
seed(0)
set_seed(0)
seed_value= 0
Neural networks in practice#
There are many practical courses on training neural nets.
We’ll use PyTorch in these examples and the labs.
Focus on key design decisions, evaluation, and regularization
Running example: Fashion-MNIST
28x28 pixel images of 10 classes of fashion items
Show code cell source
# Download FMINST data
mnist = oml.datasets.get_dataset(40996)
X, y, _, _ = mnist.get_data(target=mnist.default_target_attribute, dataset_format='array');
X = X.reshape(70000, 28, 28)
fmnist_classes = {0:"T-shirt/top", 1: "Trouser", 2: "Pullover", 3: "Dress", 4: "Coat", 5: "Sandal",
6: "Shirt", 7: "Sneaker", 8: "Bag", 9: "Ankle boot"}
# Take some random examples
from random import randint
fig, axes = plt.subplots(1, 5, figsize=(10, 5))
for i in range(5):
n = randint(0,70000)
axes[i].imshow(X[n], cmap=plt.cm.gray_r)
axes[i].set_xticks([])
axes[i].set_yticks([])
axes[i].set_xlabel("{}".format(fmnist_classes[y[n]]))
plt.show();

Preparing the data#
We’ll use feed-forward networks first, so we flatten the input data
Create train-test splits to evaluate the model later
Convert the data (numpy arrays) to PyTorch tensors
# Flatten images, create train-test split
X_flat = X.reshape(70000, 28 * 28)
X_train, X_test, y_train, y_test = train_test_split(X_flat, y, stratify=y)
# Convert numpy arrays to PyTorch tensors with correct types
X_train_tensor = torch.tensor(X_train, dtype=torch.float32)
y_train_tensor = torch.tensor(y_train, dtype=torch.long)
X_test_tensor = torch.tensor(X_test, dtype=torch.float32)
y_test_tensor = torch.tensor(y_test, dtype=torch.long)
Create data loaders to return data in batches
import torch
from torch.utils.data import DataLoader, TensorDataset
# Create PyTorch datasets
train_dataset = TensorDataset(X_train_tensor, y_train_tensor)
test_dataset = TensorDataset(X_test_tensor, y_test_tensor)
# Create DataLoaders
batch_size = 64
train_loader = DataLoader(train_dataset, batch_size=batch_size, shuffle=True)
test_loader = DataLoader(test_dataset, batch_size=batch_size, shuffle=False)
import torch
from sklearn.model_selection import train_test_split
from torch.utils.data import DataLoader, TensorDataset
# Flatten images, create train-test split
X_flat = X.reshape(70000, 28 * 28)
X_train, X_test, y_train, y_test = train_test_split(X_flat, y, stratify=y)
# Convert numpy arrays to PyTorch tensors with correct types
X_train_tensor = torch.tensor(X_train, dtype=torch.float32)
y_train_tensor = torch.tensor(y_train, dtype=torch.long)
X_test_tensor = torch.tensor(X_test, dtype=torch.float32)
y_test_tensor = torch.tensor(y_test, dtype=torch.long)
# Create PyTorch datasets
train_dataset = TensorDataset(X_train_tensor, y_train_tensor)
test_dataset = TensorDataset(X_test_tensor, y_test_tensor)
# Create DataLoaders
batch_size = 32
train_loader = DataLoader(train_dataset, batch_size=batch_size, num_workers=4, shuffle=True, pin_memory=False)
test_loader = DataLoader(test_dataset, batch_size=batch_size, num_workers=4, shuffle=False, pin_memory=False)
Building the network#
PyTorch has a Sequential and Functional API. We’ll use the Sequential API first.
Input layer: a flat vector of 28*28 = 784 nodes
We’ll see how to properly deal with images later
Two dense (Linear) hidden layers: 512 nodes each, ReLU activation
Output layer: 10 nodes (for 10 classes)
SoftMax not needed, it will be done in the loss function
import torch.nn as nn
model = nn.Sequential(
nn.Linear(28 * 28, 512), # Layer 1: 28*28 inputs to 512 output nodes
nn.ReLU(),
nn.Linear(512, 512), # Layer 2: 512 inputs to 512 output nodes
nn.ReLU(),
nn.Linear(512, 10), # Layer 3: 512 inputs to output nodes
)
In the Functional API, the same network looks like this
import torch.nn.functional as F
class NeuralNetwork(nn.Module): # Class that defines your model
def __init__(self):
super(NeuralNetwork, self).__init__() # Components defined in __init__
self.fc1 = nn.Linear(28 * 28, 512) # Fully connected layers
self.fc2 = nn.Linear(512, 512)
self.fc3 = nn.Linear(512, 10)
def forward(self, x): # Forward pass and structure of the network
x = F.relu(self.fc1(x)) # Layer 1: Input to FC1, then through ReLU
x = F.relu(self.fc2(x)) # Layer 2: Then though FC2, then ReLU
x = self.fc3(x) # Layer 3: Then though FC3, then SoftMax
return x # Return output
model = NeuralNetwork()
import torch.nn as nn
import torch.nn.functional as F
class NeuralNetwork(nn.Module): # Class that defines your model
def __init__(self):
super(NeuralNetwork, self).__init__() # Main components often defined in __init__
self.fc1 = nn.Linear(28 * 28, 512) # Fully connected layers
self.fc2 = nn.Linear(512, 512)
self.fc3 = nn.Linear(512, 10)
def forward(self, x): # Forward pass and structure of the network
x = F.relu(self.fc1(x)) # Layer 1: Input to FC1, then through ReLU
x = F.relu(self.fc2(x)) # Layer 2: Then though FC2, then ReLU
x = self.fc3(x) # Layer 3: Then though FC3, then SoftMax
return x # Return output
model = NeuralNetwork()
Choosing loss, optimizer, metrics#
Loss function: Cross-entropy (log loss) for multi-class classification
Optimizer: Any of the optimizers we discussed before. RMSprop/Adam usually work well.
Metrics: To monitor performance during training and testing, e.g. accuracy
import torch.optim as optim
import torchmetrics
# Loss function with label smoothing. Also applies softmax internally
criterion = nn.CrossEntropyLoss(label_smoothing=0.01)
# Optimizer. Note that we pass the model parameters at creation time.
optimizer = optim.RMSprop(model.parameters(), lr=0.001, momentum=0.0)
# Accuracy metric
accuracy_metric = torchmetrics.Accuracy(task="multiclass", num_classes=10)
import torch.optim as optim
import torchmetrics
cross_entropy = nn.CrossEntropyLoss(label_smoothing=0.01)
optimizer = optim.RMSprop(model.parameters(), lr=0.001, momentum=0.0)
accuracy_metric = torchmetrics.Accuracy(task="multiclass", num_classes=10)
Training on GPU#
The
deviceis where the training is done. It’s “cpu” by default.The model, tensors, and metric must all be moved to the same device.
if torch.cuda.is_available(): # For CUDA based systems
device = torch.device("cuda")
if torch.backends.mps.is_available(): # For MPS (M1-M4 Mac) based systems
device = torch.device("mps")
print(f"Used device: {device}")
# Move models and metrics to `device`
model.to(device)
accuracy_metric = accuracy_metric.to(device)
# Move batches one at a time (GPUs have limited memory)
for X_batch, y_batch in train_loader:
X_batch, y_batch = X_batch.to(device), y_batch.to(device)
if torch.cuda.is_available(): # For CUDA based systems
device = torch.device("cuda")
if torch.backends.mps.is_available(): # For MPS (M1-M4 Mac) based systems
device = torch.device("mps")
print(f"Used device: {device}")
# Move everything to `device`
model.to(device)
accuracy_metric = accuracy_metric.to(device)
for X_batch, y_batch in train_loader:
X_batch, y_batch = X_batch.to(device), y_batch.to(device)
break
Used device: mps
Training loop#
In pure PyTorch, you have to write the training loop yourself (as well as any code to print out progress)
for epoch in range(10):
for X_batch, y_batch in train_loader:
X_batch, y_batch = X_batch.to(device), y_batch.to(device) # to GPU
# Forward pass + loss calculation
outputs = model(X_batch)
loss = cross_entropy(outputs, y_batch)
# Backward pass
optimizer.zero_grad() # Reset gradients (otherwise they accumulate)
loss.backward() # Backprop. Computes all gradients
optimizer.step() # Uses gradients to update weigths
num_epochs = 5
for epoch in range(5):
total_loss, correct, total = 0, 0, 0
for X_batch, y_batch in train_loader:
X_batch, y_batch = X_batch.to(device), y_batch.to(device) # to GPU
# Forward pass + loss calculation
outputs = model(X_batch)
loss = cross_entropy(outputs, y_batch)
# Backward pass: zero gradients, do backprop, do optimizer step
optimizer.zero_grad()
loss.backward()
optimizer.step()
# Compute training metrics
total_loss += loss.item()
correct += accuracy_metric(outputs, y_batch).item() * y_batch.size(0)
total += y_batch.size(0)
# Compute epoch metrics
avg_loss = total_loss / len(train_loader)
avg_acc = correct / total
print(f"Epoch [{epoch+1}/{num_epochs}], Loss: {avg_loss:.4f}, Accuracy: {avg_acc:.4f}")
Epoch [1/5], Loss: 2.3524, Accuracy: 0.7495
Epoch [2/5], Loss: 0.5531, Accuracy: 0.8259
Epoch [3/5], Loss: 0.5102, Accuracy: 0.8408
Epoch [4/5], Loss: 0.4897, Accuracy: 0.8493
Epoch [5/5], Loss: 0.4758, Accuracy: 0.8550
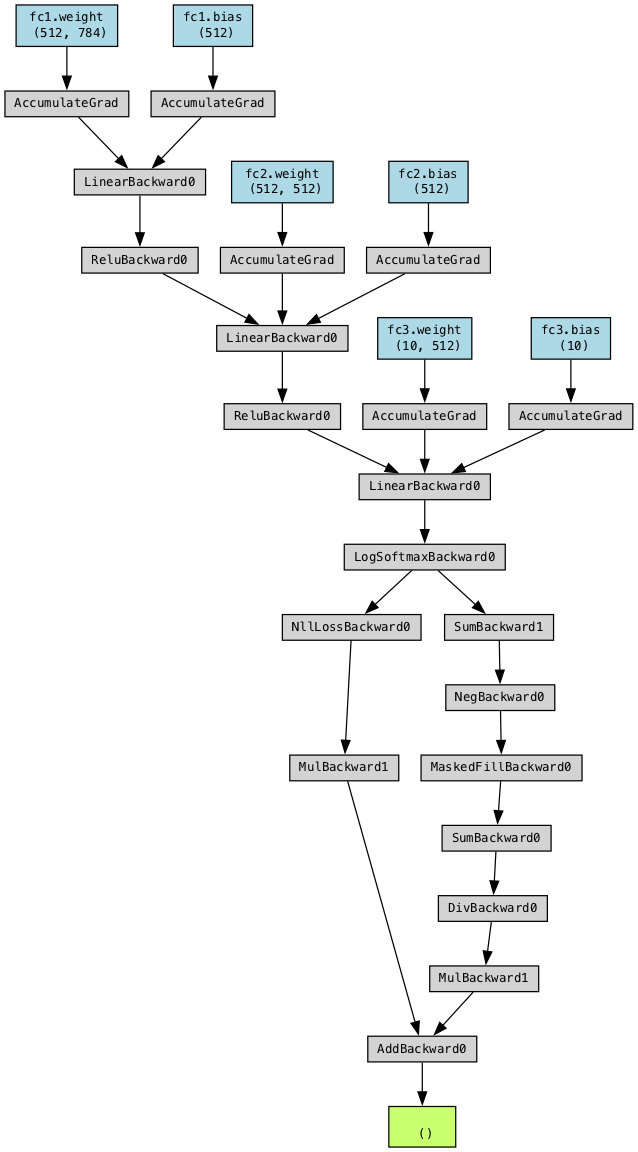
loss.backward()#
Every time you perform a forward pass, PyTorch dynamically constructs a computational graph
This graph tracks tensors and operations involved in computing gradients (see next slide)
The
lossreturned is a tensor, and every tensor is part of the computational graphWhen you call
.backward()onloss, PyTorch traverses this graph in reverse to compute all gradientsThis process is called automatic differentiation
Stores intermediate values so no gradient component is calculated twice
When
backward()completes, the computational graph is discarded by default to free memory
Computational graph for our model (Loss in green, weights/biases in blue)

from torchviz import make_dot
from IPython.display import Image
toy_X = X_train_tensor[0].to(device)
toy_y = y_train_tensor[0].to(device)
toy_out = model(toy_X)
loss = cross_entropy(toy_out, toy_y)
graph = make_dot(loss, params=dict(model.named_parameters()))
graph.render("computational_graph", format="png", view=True)
Image("computational_graph.png")
In PyTorch Lightning#
A high-level framework built on PyTorch that simplifies deep learning model training
Same code, but extend
pl.LightingModuleinstead ofnn.ModuleHas a number of predefined functions. For instance:
class NeuralNetwork(pl.LightningModule):
def __init__(self):
pass # Initialize model
def forward(self, x):
pass # Forward pass, return output tensor
def configure_optimizers(self):
pass # Configure optimizer (e.g. Adam)
def training_step(self, batch, batch_idx):
pass # Return loss tensor
def validation_step(self, batch, batch_idx):
pass # Return loss tensor
def test_step(self, batch, batch_idx):
pass # Return loss tensor
Our entire example now becomes:
import pytorch_lightning as pl
class NeuralNetwork(pl.LightningModule):
def __init__(self):
super(LitNeuralNetwork, self).__init__()
self.fc1 = nn.Linear(28 * 28, 512)
self.fc2 = nn.Linear(512, 512)
self.fc3 = nn.Linear(512, 10)
self.criterion = nn.CrossEntropyLoss(label_smoothing=0.01)
self.accuracy = torchmetrics.Accuracy(task="multiclass", num_classes=10)
def forward(self, x):
x = F.relu(self.fc1(x))
x = F.relu(self.fc2(x))
return self.fc3(x)
def training_step(self, batch, batch_idx):
X_batch, y_batch = batch
outputs = self(X_batch)
return self.criterion(outputs, y_batch)
def configure_optimizers(self):
return optim.RMSprop(self.parameters(), lr=0.001, momentum=0.0)
model = NeuralNetwork()
import pytorch_lightning as pl
class NeuralNetwork(pl.LightningModule):
def __init__(self):
super(NeuralNetwork, self).__init__()
self.fc1 = nn.Linear(28 * 28, 512)
self.fc2 = nn.Linear(512, 512)
self.fc3 = nn.Linear(512, 10)
self.criterion = nn.CrossEntropyLoss(label_smoothing=0.01)
self.accuracy = torchmetrics.Accuracy(task="multiclass", num_classes=10)
def forward(self, x):
x = x.view(x.size(0), -1) # Flatten the image
x = F.relu(self.fc1(x))
x = F.relu(self.fc2(x))
return self.fc3(x)
def training_step(self, batch, batch_idx):
X_batch, y_batch = batch
outputs = self(X_batch) # Logits (raw outputs)
loss = self.criterion(outputs, y_batch) # Loss
preds = torch.argmax(outputs, dim=1) # Predictions
acc = self.accuracy(preds, y_batch)
self.log("train_loss", loss, sync_dist=True)
self.log("train_acc", acc, sync_dist=True)
return loss
def validation_step(self, batch, batch_idx):
X_batch, y_batch = batch
outputs = self(X_batch)
loss = self.criterion(outputs, y_batch)
preds = torch.argmax(outputs, dim=1)
acc = self.accuracy(preds, y_batch)
self.log("val_loss", loss, sync_dist=True, on_epoch=True)
self.log("val_acc", acc, sync_dist=True, on_epoch=True)
return loss
def on_train_epoch_end(self):
avg_loss = self.trainer.callback_metrics["train_loss"].item()
avg_acc = self.trainer.callback_metrics["train_acc"].item()
print(f"Epoch {self.trainer.current_epoch+1}: Loss = {avg_loss:.4f}, Accuracy = {avg_acc:.4f}")
def configure_optimizers(self):
return optim.RMSprop(self.parameters(), lr=0.001, momentum=0.0)
model = NeuralNetwork()
We can also get a nice model summary
Lots of parameters (weights and biases) to learn!
hidden layer 1 : (28 * 28 + 1) * 512 = 401920
hidden layer 2 : (512 + 1) * 512 = 262656
output layer: (512 + 1) * 10 = 5130
ModelSummary(pl_model, max_depth=2)
Show code cell source
from pytorch_lightning.utilities.model_summary import ModelSummary
ModelSummary(model, max_depth=2)
| Name | Type | Params | Mode
---------------------------------------------------------
0 | fc1 | Linear | 401 K | train
1 | fc2 | Linear | 262 K | train
2 | fc3 | Linear | 5.1 K | train
3 | criterion | CrossEntropyLoss | 0 | train
4 | accuracy | MulticlassAccuracy | 0 | train
---------------------------------------------------------
669 K Trainable params
0 Non-trainable params
669 K Total params
2.679 Total estimated model params size (MB)
5 Modules in train mode
0 Modules in eval mode
Training#
To log results while training, we can extend the training methods:
def training_step(self, batch, batch_idx):
X_batch, y_batch = batch
outputs = self(X_batch) # Logits (raw outputs)
loss = self.criterion(outputs, y_batch) # Loss
preds = torch.argmax(outputs, dim=1) # Predictions
acc = self.accuracy(preds, y_batch) # Metric
self.log("train_loss", loss) # self.log is the default
self.log("train_acc", acc) # TensorBoard logger
return loss
def on_train_epoch_end(self): # Runs at the end of every epoch
avg_loss = self.trainer.callback_metrics["train_loss"].item()
avg_acc = self.trainer.callback_metrics["train_acc"].item()
print(f"Epoch {self.trainer.current_epoch}: Loss = {avg_loss:.4f}, Train accuracy = {avg_acc:.4f}")
We also need to implement the validation steps if we want validation scores
Identical to
training_stepexcept for the logging
def validation_step(self, batch, batch_idx):
X_batch, y_batch = batch
outputs = self(X_batch)
loss = self.criterion(outputs, y_batch)
preds = torch.argmax(outputs, dim=1)
acc = self.accuracy(preds, y_batch)
self.log("val_loss", loss, on_epoch=True)
self.log("val_acc", acc, on_epoch=True)
return loss
Lightning Trainer#
For training, we can now create a trainer and fit it. This will also automatically move everything to GPU.
trainer = pl.Trainer(max_epochs=3, accelerator="gpu") # Or 'cpu'
trainer.fit(model, train_loader)
import logging # Cleaner output
logging.getLogger("pytorch_lightning").setLevel(logging.ERROR)
accelerator = "cpu"
if torch.backends.mps.is_available():
accelerator = "mps"
if torch.cuda.is_available():
accelerator = "gpu"
trainer = pl.Trainer(max_epochs=3, accelerator=accelerator)
trainer.fit(model, train_loader)
Epoch 1: Loss = 0.6928, Accuracy = 0.8000
Epoch 2: Loss = 0.3986, Accuracy = 0.9000
Epoch 3: Loss = 0.3572, Accuracy = 0.9000
Choosing training hyperparameters#
Number of epochs: enough to allow convergence
Too much: model starts overfitting (or levels off and just wastes time)
Batch size: small batches (e.g. 32, 64,… samples) often preferred
‘Noisy’ training data makes overfitting less likely
Large batches generalize less well (‘generalization gap’)
Requires less memory (especially in GPUs)
Large batches do speed up training, may converge in fewer epochs
Batch size interacts with learning rate
Instead of shrinking the learning rate you can increase batch size
# Same model, more silent
class NeuralNetwork(pl.LightningModule):
def __init__(self):
super(NeuralNetwork, self).__init__()
self.fc1 = nn.Linear(28 * 28, 512)
self.fc2 = nn.Linear(512, 512)
self.fc3 = nn.Linear(512, 10)
self.criterion = nn.CrossEntropyLoss(label_smoothing=0.01)
self.accuracy = torchmetrics.Accuracy(task="multiclass", num_classes=10)
def forward(self, x):
x = x.view(x.size(0), -1) # Flatten the image
x = F.relu(self.fc1(x))
x = F.relu(self.fc2(x))
return self.fc3(x)
def training_step(self, batch, batch_idx):
X_batch, y_batch = batch
outputs = self(X_batch) # Logits (raw outputs)
loss = self.criterion(outputs, y_batch) # Loss
preds = torch.argmax(outputs, dim=1) # Predictions
acc = self.accuracy(preds, y_batch)
self.log("train_loss", loss, sync_dist=True)
self.log("train_acc", acc, sync_dist=True)
return loss
def validation_step(self, batch, batch_idx):
X_batch, y_batch = batch
outputs = self(X_batch)
loss = self.criterion(outputs, y_batch)
preds = torch.argmax(outputs, dim=1)
acc = self.accuracy(preds, y_batch)
self.log("val_loss", loss, sync_dist=True, on_epoch=True)
self.log("val_acc", acc, sync_dist=True, on_epoch=True)
return loss
def on_train_epoch_end(self):
avg_loss = self.trainer.callback_metrics["train_loss"].item()
avg_acc = self.trainer.callback_metrics["train_acc"].item()
def on_validation_epoch_end(self):
avg_loss = self.trainer.callback_metrics["val_loss"].item()
avg_acc = self.trainer.callback_metrics["val_acc"].item()
def configure_optimizers(self):
return optim.RMSprop(self.parameters(), lr=0.001, momentum=0.0)
model = NeuralNetwork()
from pytorch_lightning.callbacks import Callback
from IPython.display import clear_output
# First try. Not very efficient yet.
class TrainingBatchPlotCallback(Callback):
def __init__(self, update_interval=100, smoothing=100):
"""
Callback to plot training and validation progress.
Args:
update_interval (int): Number of batches between updates.
smoothing (int): Window size for moving average.
"""
super().__init__()
self.update_interval = update_interval
self.smoothing = smoothing
self.losses = []
self.acc = []
self.val_losses = []
self.val_acc = []
self.val_x_positions = [] # Store x positions for validation scores
self.max_acc = 0
self.global_step = 0 # Tracks processed training batches
self.epoch_count = 0 # Tracks processed epochs
self.steps_per_epoch = 0 # Updated dynamically to track num_batches per epoch
def moving_average(self, values, window):
"""Computes a simple moving average for smoothing."""
if len(values) < window:
return np.convolve(values, np.ones(len(values)) / len(values), mode="valid")
return np.convolve(values, np.ones(window) / window, mode="valid")
def on_train_batch_end(self, trainer, pl_module, outputs, batch, batch_idx):
"""Updates training loss and accuracy every `update_interval` batches."""
self.global_step += 1
logs = trainer.callback_metrics
# Store training loss and accuracy
if "train_loss" in logs:
self.losses.append(logs["train_loss"].item())
if "train_acc" in logs:
self.acc.append(logs["train_acc"].item())
# Update plot every `update_interval` training batches
if self.global_step % self.update_interval == 0:
self.plot_progress()
def on_validation_epoch_end(self, trainer, pl_module):
"""Updates validation loss and accuracy at the end of each epoch, scaling x-axis correctly."""
logs = trainer.callback_metrics
self.epoch_count += 1
if self.steps_per_epoch == 0:
self.steps_per_epoch = self.global_step
self.val_x_positions.append(self.global_step)
# Retrieve validation metrics from trainer
if "val_loss" in logs:
self.val_losses.append(logs["val_loss"].item())
if "val_acc" in logs:
val_acc = logs["val_acc"].item()
self.val_acc.append(val_acc)
self.max_acc = max(self.max_acc, val_acc)
def on_train_end(self, trainer, pl_module):
"""Ensures the final validation curve is plotted after the last epoch."""
self.plot_progress()
def plot_progress(self):
"""Plots training and validation metrics with moving averages."""
clear_output(wait=True)
steps = np.arange(1, len(self.losses) + 1)
plt.figure(figsize=(8, 4))
plt.ylim(0, 1) # Constrain y-axis between 0 and 1
# Training curves
plt.plot(steps / self.steps_per_epoch, self.losses, lw=0.2, alpha=0.3, c="b", linestyle="-")
plt.plot(steps / self.steps_per_epoch, self.acc, lw=0.2, alpha=0.3, c="r", linestyle="-")
# Moving average (thicker line)
if len(self.losses) >= self.smoothing:
plt.plot(steps[self.smoothing - 1:] / self.steps_per_epoch, self.moving_average(self.losses, self.smoothing),
lw=1, c="b", linestyle="-", label="Train Loss")
if len(self.acc) >= self.smoothing:
plt.plot(steps[self.smoothing - 1:] / self.steps_per_epoch, self.moving_average(self.acc, self.smoothing),
lw=1, c="r", linestyle="-", label="Train Acc")
# Validation curves (scaled to correct x-positions)
if self.val_losses:
plt.plot(np.array(self.val_x_positions) / self.steps_per_epoch, self.val_losses, c="b", linestyle=":", label="Val Loss", lw=2)
if self.val_acc:
plt.plot(np.array(self.val_x_positions) / self.steps_per_epoch, self.val_acc, c="r", linestyle=":", label="Val Acc", lw=2)
plt.xlabel("Epochs", fontsize=12)
plt.ylabel("Loss/Accuracy", fontsize=12)
plt.title(f"Training Progress (Step {self.global_step}, Epoch {self.epoch_count}, Max Val Acc {self.max_acc:.4f})", fontsize=12)
plt.xticks(fontsize=10)
plt.yticks(fontsize=10)
plt.legend(fontsize=10)
plt.show()
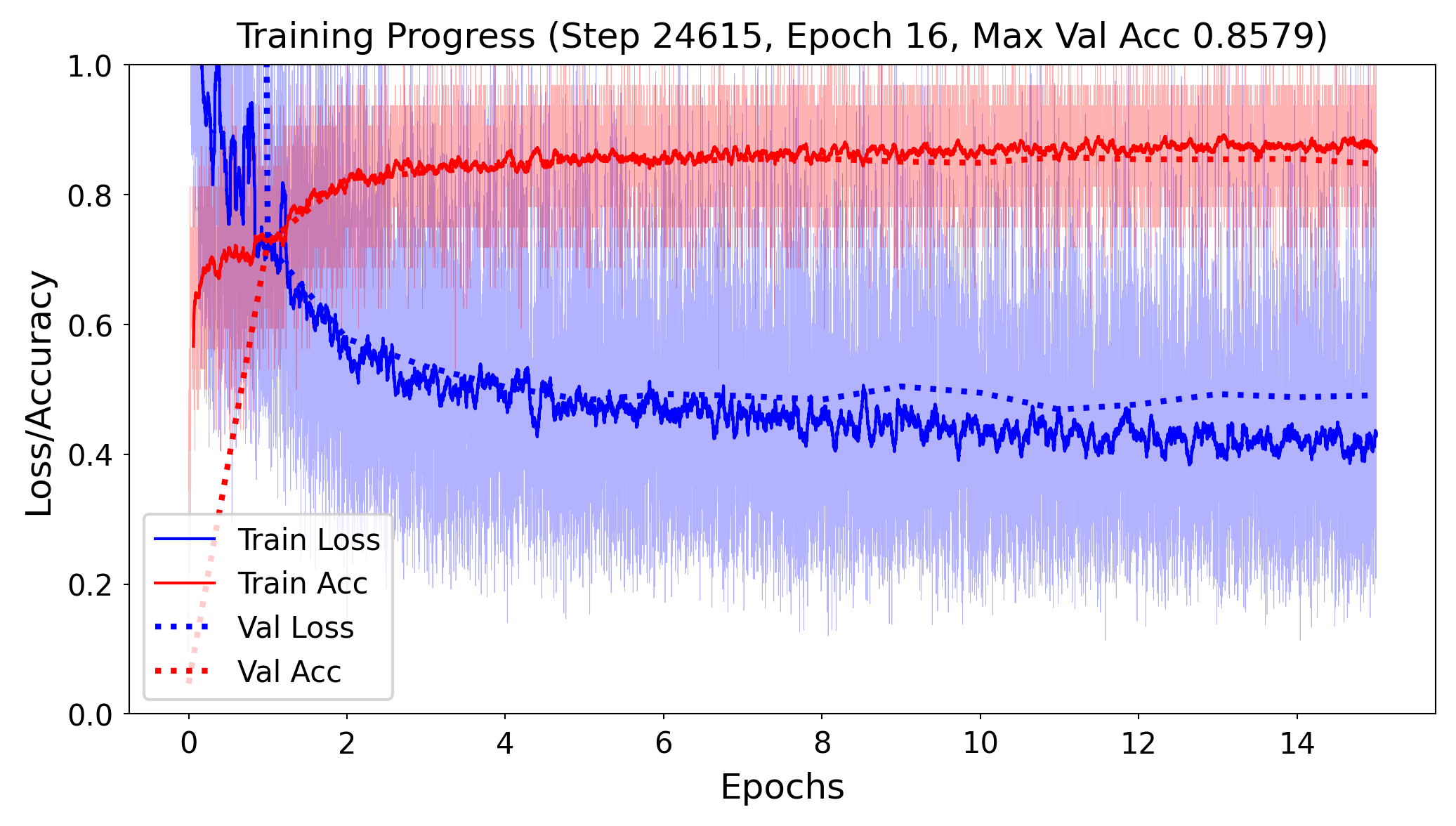
Model selection#
Train the neural net and track the loss after every iteration on a validation set
You can add a callback to the fit version to get info on every epoch
Best model after a few epochs, then starts overfitting
trainer = pl.Trainer(
max_epochs=15,
enable_progress_bar=False,
accelerator=accelerator,
callbacks=[TrainingBatchPlotCallback()] # Attach the callback
)
trainer.fit(model, train_loader, test_loader)
Early stopping#
Stop training when the validation loss (or validation accuracy) no longer improves
Loss can be bumpy: use a moving average or wait for \(k\) steps without improvement
# Define early stopping callback
early_stopping = EarlyStopping(
monitor="val_loss", mode="min", # minimize validation loss
patience=3) # Number of epochs with no improvement before stopping
# Update the Trainer to include early stopping as a callback
trainer = pl.Trainer(
max_epochs=10, accelerator=accelerator,
callbacks=[TrainingPlotCallback(), early_stopping] # Attach the callbacks
)
Regularization and memorization capacity#
The number of learnable parameters is called the model capacity
A model with more parameters has a higher memorization capacity
Too high capacity causes overfitting, too low causes underfitting
In the extreme, the training set can be ‘memorized’ in the weights
Smaller models are forced it to learn a compressed representation that generalizes better
Find the sweet spot: e.g. start with few parameters, increase until overfitting stars.
Example: 256 nodes in first layer, 32 nodes in second layer, similar performance
Avoid bottlenecks: layers so small that information is lost
self.fc1 = nn.Linear(28 * 28, 256)
self.fc2 = nn.Linear(256, 32)
self.fc3 = nn.Linear(32, 10)
Weight regularization (weight decay)#
We can also add weight regularization to our loss function (or invent your own)
L1 regularization: leads to sparse networks with many weights that are 0
L2 regularization: leads to many very small weights
def training_step(self, batch, batch_idx):
X_batch, y_batch = batch
outputs = self(X_batch)
loss = self.criterion(outputs, y_batch)
l1_lambda = 1e-5 # L1 Regularization
l1_loss = sum(p.abs().sum() for p in self.parameters())
l2_lambda = 1e-4 # L2 Regularization
l2_loss = sum((p ** 2).sum() for p in self.parameters())
return loss + l2_lambda * l2_loss # Using L2 only
Alternative: set weight_decay in the optimizer (only for L2 loss)
def configure_optimizers(self):
return optim.RMSprop(self.parameters(), lr=0.001, momentum=0.0, weight_decay=1e-4)
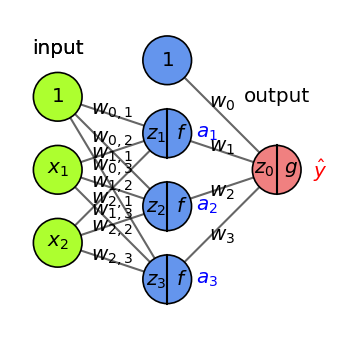
Dropout#
Every iteration, randomly set a number of activations \(a_i\) to 0
Dropout rate : fraction of the outputs that are zeroed-out (e.g. 0.1 - 0.5)
Use higher dropout rates for deeper networks
Use higher dropout in early layers, lower dropout later
Early layers are usually larger, deeper layers need stability
Idea: break up accidental non-significant learned patterns
At test time, nothing is dropped out, but the output values are scaled down by the dropout rate
Balances out that more units are active than during training
Show code cell source
fig = plt.figure(figsize=(3*fig_scale, 3*fig_scale))
ax = fig.gca()
draw_neural_net(ax, [2, 3, 1], draw_bias=True, labels=True,
show_activations=True, activation=True)
Dropout layers#
Dropout is usually implemented as a special layer
def __init__(self):
super(NeuralNetwork, self).__init__()
self.fc1 = nn.Linear(28 * 28, 512)
self.dropout1 = nn.Dropout(p=0.2) # 20% dropout
self.fc2 = nn.Linear(512, 512)
self.dropout2 = nn.Dropout(p=0.1) # 10% dropout
self.fc3 = nn.Linear(512, 10)
def forward(self, x):
x = F.relu(self.fc1(x))
x = self.dropout1(x) # Apply dropout
x = F.relu(self.fc2(x))
x = self.dropout2(x) # Apply dropout
return self.fc3(x)
Batch Normalization#
We’ve seen that scaling the input is important, but what if layer activations become very large?
Same problems, starting deeper in the network
Batch normalization: normalize the activations of the previous layer within each batch
Within a batch, set the mean activation close to 0 and the standard deviation close to 1
Across badges, use exponential moving average of batch-wise mean and variance
Allows deeper networks less prone to vanishing or exploding gradients
BatchNorm layers#
Batch normalization is also usually implemented as a special layer
def __init__(self):
super(NeuralNetwork, self).__init__()
self.fc1 = nn.Linear(28 * 28, 512)
self.bn1 = nn.BatchNorm1d(512) # Batch normalization after first layer
self.fc2 = nn.Linear(512, 265)
self.bn2 = nn.BatchNorm1d(265) # Batch normalization after second layer
self.fc3 = nn.Linear(265, 10)
def forward(self, x):
x = x.view(x.size(0), -1) # Flatten the image
x = F.relu(self.bn1(self.fc1(x))) # Apply batch norm after linear layer
x = F.relu(self.bn2(self.fc2(x))) # Apply batch norm after second layer
return self.fc3(x)
New model#
class NeuralNetwork(pl.LightningModule):
def __init__(self):
super(NeuralNetwork, self).__init__()
self.fc1 = nn.Linear(28 * 28, 265)
self.bn1 = nn.BatchNorm1d(265)
self.dropout1 = nn.Dropout(0.5)
self.fc2 = nn.Linear(265, 64)
self.bn2 = nn.BatchNorm1d(64)
self.dropout2 = nn.Dropout(0.5)
self.fc3 = nn.Linear(64, 32)
self.bn3 = nn.BatchNorm1d(32)
self.dropout3 = nn.Dropout(0.5)
self.fc4 = nn.Linear(32, 10)
self.criterion = nn.CrossEntropyLoss(label_smoothing=0.01)
self.accuracy = torchmetrics.Accuracy(task="multiclass", num_classes=10)
def forward(self, x):
x = F.relu(self.bn1(self.fc1(x)))
x = self.dropout1(x)
x = F.relu(self.bn2(self.fc2(x)))
x = self.dropout2(x)
x = F.relu(self.bn3(self.fc3(x)))
x = self.dropout3(x)
x = self.fc4(x)
return x
New model (Sequential API)#
model = nn.Sequential(
nn.Linear(28 * 28, 265),
nn.BatchNorm1d(265),
nn.ReLU(),
nn.Dropout(0.5),
nn.Linear(265, 64),
nn.BatchNorm1d(64),
nn.ReLU(),
nn.Dropout(0.5),
nn.Linear(64, 32),
nn.BatchNorm1d(32),
nn.ReLU(),
nn.Dropout(0.5),
nn.Linear(32, 10))
Our model now performs better and is improving.
class NeuralNetwork(pl.LightningModule):
def __init__(self):
super(NeuralNetwork, self).__init__()
self.fc1 = nn.Linear(28 * 28, 265)
self.bn1 = nn.BatchNorm1d(265)
self.dropout1 = nn.Dropout(0.5)
self.fc2 = nn.Linear(265, 64)
self.bn2 = nn.BatchNorm1d(64)
self.dropout2 = nn.Dropout(0.5)
self.fc3 = nn.Linear(64, 32)
self.bn3 = nn.BatchNorm1d(32)
self.dropout3 = nn.Dropout(0.5)
self.fc4 = nn.Linear(32, 10)
self.accuracy = torchmetrics.Accuracy(task="multiclass", num_classes=10)
self.criterion = nn.CrossEntropyLoss(label_smoothing=0.01)
def forward(self, x):
x = F.relu(self.bn1(self.fc1(x)))
x = self.dropout1(x)
x = F.relu(self.bn2(self.fc2(x)))
x = self.dropout2(x)
x = F.relu(self.bn3(self.fc3(x)))
x = self.dropout3(x)
x = self.fc4(x)
return x
def training_step(self, batch, batch_idx):
X_batch, y_batch = batch
outputs = self(X_batch) # Logits (raw outputs)
loss = self.criterion(outputs, y_batch) # Loss
preds = torch.argmax(outputs, dim=1) # Predictions
acc = self.accuracy(preds, y_batch)
self.log("train_loss", loss, sync_dist=True)
self.log("train_acc", acc, sync_dist=True)
return loss
def validation_step(self, batch, batch_idx):
X_batch, y_batch = batch
outputs = self(X_batch)
loss = self.criterion(outputs, y_batch)
preds = torch.argmax(outputs, dim=1)
acc = self.accuracy(preds, y_batch)
self.log("val_loss", loss, sync_dist=True, on_epoch=True)
self.log("val_acc", acc, sync_dist=True, on_epoch=True)
return loss
def on_train_epoch_end(self):
avg_loss = self.trainer.callback_metrics["train_loss"].item()
avg_acc = self.trainer.callback_metrics["train_acc"].item()
def on_validation_epoch_end(self):
avg_loss = self.trainer.callback_metrics["val_loss"].item()
avg_acc = self.trainer.callback_metrics["val_acc"].item()
def configure_optimizers(self):
return optim.RMSprop(self.parameters(), lr=0.001, momentum=0.0)
model = NeuralNetwork()
trainer = pl.Trainer(
max_epochs=10,
enable_progress_bar=False,
accelerator=accelerator,
callbacks=[TrainingBatchPlotCallback()] # Attach the callback
)
trainer.fit(model, train_loader, test_loader)
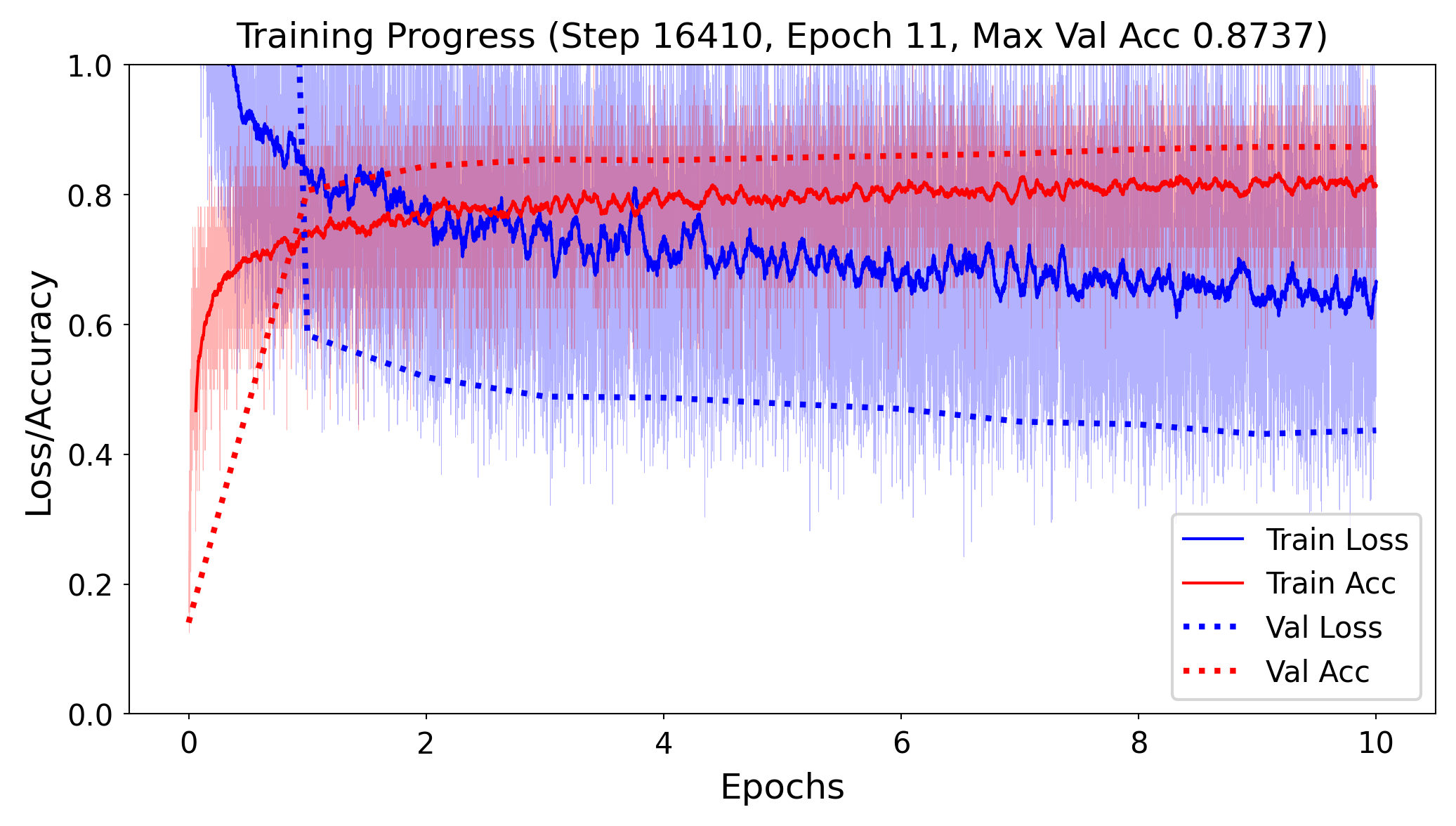
Other logging tools#
There is a lot more tooling to help you build good models
E.g. TensorBoard is easy to integrate and offers a convenient dashboard
logger = pl.loggers.TensorBoardLogger("logs/", name="my_experiment")
trainer = pl.Trainer(max_epochs=2, logger=logger)
trainer.fit(lit_model, trainloader)

Summary#
Neural architectures
Training neural nets
Forward pass: Tensor operations
Backward pass: Backpropagation
Neural network design:
Activation functions
Weight initialization
Optimizers
Neural networks in practice
Model selection
Early stopping
Memorization capacity and information bottleneck
L1/L2 regularization
Dropout
Batch normalization