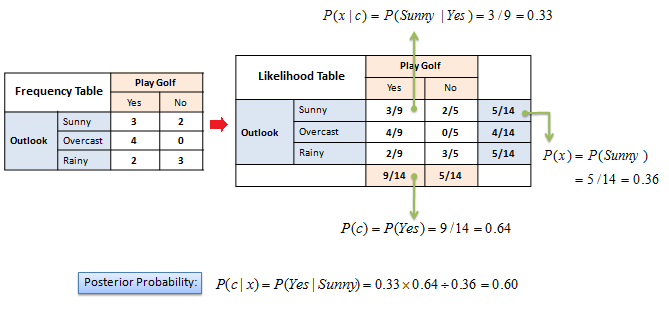
 XKCD, Randall Monroe
XKCD, Randall Monroe
Bayes' rule¶
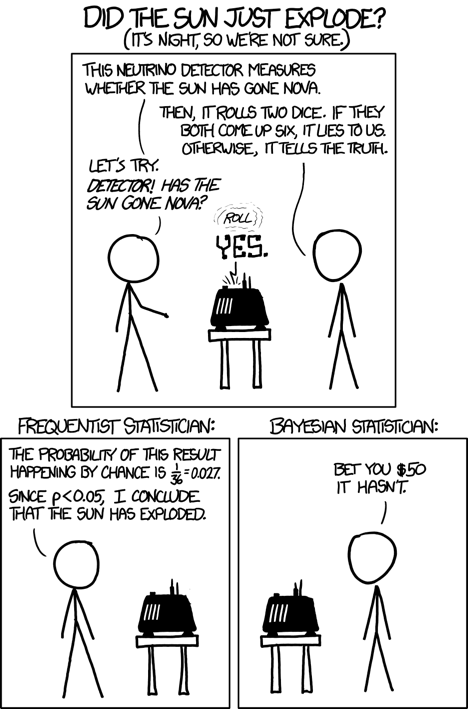
Rule for updating the probability of a hypothesis $c$ given data $x$
$P(c|x)$ is the posterior probability of class $c$ given data $x$.
$P(c)$ is the prior probability of class $c$: what you believed before you saw the data $x$
$P(x|c)$ is the likelihood of data point $x$ given that the class is $c$ (computed from your dataset)
$P(x)$ is the prior probability of the data (marginal likelihood): the likelihood of the data $x$ under any circumstance (no matter what the class is)
Example: exploding sun¶
- Let's compute the probability that the sun has exploded
- Prior $P(exploded)$: the sun has an estimated lifespan of 10 billion years, $P(exploded) = \frac{1}{4.38 x 10^{13}}$
- Likelihood that detector lies: $P(lie)= \frac{1}{36}$
- The one positive observation of the detector increases the probability
Example: COVID test¶
- What is the probability of having COVID-19 if a 96% accurate test returns positive? Assume a false positive rate of 4%
- Prior $P(C): 0.015$ (117M cases, 7.9B people)
- $P(TP)=P(pos|C)=0.96$, and $P(FP)=(pos|notC)=0.04$
- If test is positive, prior becomes $P(C)=0.268$. 2nd positive test: $P(C|pos)=0.9$
Bayesian models¶
- Learn the joint distribution $P(x,y)=P(x|y)P(y)$.
- Assumes that the data is Gaussian distributed (!)
- With every input $x$ you get $P(y|x)$, hence a mean and standard deviation for $y$ (blue)
- For every desired output $y$ you get $P(x|y)$, hence you can sample new points $x$ (red)
- Easily updatable with new data using Bayes' rule ('turning the crank')
- Previous posterior $P(y|x)$ becomes new prior $P(y)$
interactive(children=(FloatSlider(value=0.0, description='x', max=3.0, min=-3.0, step=0.5), FloatSlider(value=…
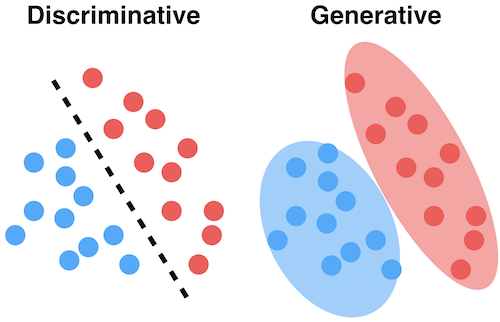
Generative models¶
- The joint distribution represents the training data for a particular output (e.g. a class)
- You can sample a new point $\textbf{x}$ with high predicted likelihood $P(x,c)$: that new point will be very similar to the training points
- Generate new (likely) points according to the same distribution: generative model
- Generate examples that are fake but corresponding to a desired output
- Generative neural networks (e.g. GANs) can do this very accurately for text, images, ...
Naive Bayes¶
- Predict the probability that a point belongs to a certain class, using Bayes' Theorem
- Problem: since $\textbf{x}$ is a vector, computing $P(\textbf{x}|c)$ can be very complex
- Naively assume that all features are conditionally independent from each other, in which case:
$P(\mathbf{x}|c) = P(x_1|c) \times P(x_2|c) \times ... \times P(x_n|c)$ - Very fast: only needs to extract statistics from each feature.
On numeric data¶
- We need to fit a distribution (e.g. Gaussian) over the data points
GaussianNB: Computes mean $\mu_c$ and standard deviation $\sigma_c$ of the feature values per class: $p(x=v \mid c)=\frac{1}{\sqrt{2\pi\sigma^2_c}}\,e^{ -\frac{(v-\mu_c)^2}{2\sigma^2_c} }$
We can now make predictions using Bayes' theorem: $p(c \mid \mathbf{x}) = \frac{p(\mathbf{x} \mid c) \ p(c)}{p(\mathbf{x})}$
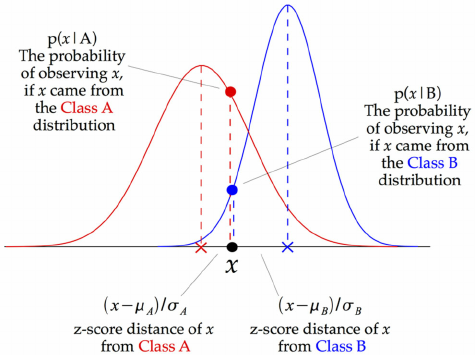
- What do the predictions of Gaussian Naive Bayes look like?
Other Naive Bayes classifiers:
- BernoulliNB
- Assumes binary data
- Feature statistics: Number of non-zero entries per class
- MultinomialNB
- Assumes count data
- Feature statistics: Average value per class
- Mostly used for text classification (bag-of-words data)
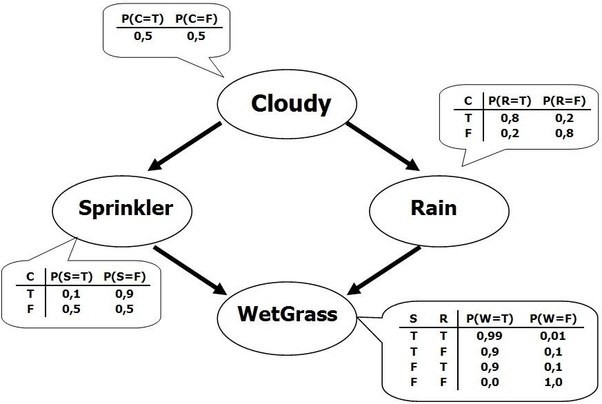
Bayesian Networks¶
- What if we know that some variables are not independent?
- A Bayesian Network is a directed acyclic graph representing variables as nodes and conditional dependencies as edges.
- If an edge $(A, B)$ connects random variables A and B, then $P(B|A)$ is a factor in the joint probability distribution. We must know $P(B|A)$ for all values of $B$ and $A$
- The graph structure can be designed manually or learned (hard!)
Gaussian processes¶
- Model the data as a Gaussian distribution, conditioned on the training points
interactive(children=(IntSlider(value=10, description='nr_points', max=20), Output()), _dom_classes=('widget-i…
Probabilistic interpretation of regression¶
Linear regression (recap):
$$y = f(\mathbf{x}_i) = \mathbf{x}_i\mathbf{w} + b $$For one input feature: $$y = w_1 \cdot x_1 + b \cdot 1$$
We can solve this via linear algebra (closed form solution): $w^{*} = (X^{T}X)^{-1} X^T Y$
w = np.linalg.solve(np.dot(X.T, X), np.dot(X.T, y))
$\mathbf{X}$ is our data matrix with a $x_0=1$ column to represent the bias $b$:
$$\mathbf{X} = \begin{bmatrix} \mathbf{x}_1^\top \\\ \mathbf{x}_2^\top \\\ \vdots \\\ \mathbf{x}_N^\top \end{bmatrix} = \begin{bmatrix} 1 & x_1 \\\ 1 & x_2 \\\ \vdots & \vdots \\\ 1 & x_N \end{bmatrix}$$Example: Olympic marathon data¶
We learned: $ y= w_1 x + w_0 = -0.013 x + 28.895$
Polynomial regression (recap)¶
We can fit a 2nd degree polynomial by using a basis expansion (adding more basis functions):
$$\mathbf{\Phi} = \left[ \mathbf{1} \quad \mathbf{x} \quad \mathbf{x}^2\right]$$Kernelized regression (recap)¶
We can also kernelize the model and learn a dual coefficient per data point
interactive(children=(IntSlider(value=4, description='poly_degree', max=8, min=1), IntSlider(value=-6, descrip…
Probabilistic interpretation¶
- These models do not give us any indication of the (un)certainty of the predictions
- Assume that the data is inherently uncertain. This can be modeled explictly by introducing a slack variable, $\epsilon_i$, known as noise.
- Assume that the noise is distributed according to a Gaussian distribution with zero mean and variance $\sigma^2$.
- That means that $y(x)$ is now a Gaussian distribution with mean $\mathbf{wx}$ and variance $\sigma^2$
We have an uncertainty predictions, but it is the same for all predictions
- You would expect to be more certain nearby your training points
interactive(children=(FloatSlider(value=0.005, description='sigma', max=0.01, min=0.001, step=0.001), Output()…
How to learn probabilities?¶
- Maximum Likelihood Estimation (MLE): Maximize $P(\textbf{X}|\textbf{w})$
- Corresponds to optimizing $\mathbf{w}$, using (log) likelihood as the loss function
- Every prediction has a mean defined by $\textbf{w}$ and Gaussian noise $$P(\textbf{X}|\textbf{w}) = \prod_{i=0}^{n} P(\mathbf{y}_i|\mathbf{x}_i;\mathbf{w}) = \prod_{i=0}^{n} \mathcal{N}(\mathbf{wx,\sigma^2 I})$$
- Maximum A Posteriori estimation (MAP): Maximize the posterior $P(\textbf{w}|\textbf{X})$
- This can be done using Bayes' rule after we choose a (Gaussian) prior $P(\textbf{w})$: $$P(\textbf{w}|\textbf{X}) = \frac{P(\textbf{X}|\textbf{w})P(\textbf{w})}{P(\textbf{X})}$$
- Bayesian approach: model the prediction $P(y|x_{test},X)$ directly
- Marginalize $w$ out: consider all possible models (some are more likely)
- If prior $P(\textbf{w})$ is Gaussian, then $P(y|x_{test},\textbf{X})$ is also Gaussian!
- A multivariate Gaussian with mean $\mu$ and covariance matrix $\Sigma$ $$P(y|x_{test},\textbf{X}) = \int_w P(y|x_{test},\textbf{w}) P(\textbf{w}|\textbf{X}) dw = \mathcal{N}(\mathbf{\mu,\Sigma})$$
Gaussian prior $P(w)$¶
In the Bayesian approach, we assume a prior (Gaussian) distribution for the parameters, $\mathbf{w} \sim \mathcal{N}(\mathbf{0}, \alpha \mathbf{I})$:
- With zero mean ($\mu$=0) and covariance matrix $\alpha \mathbf{I}$. For 2D: $ \alpha \mathbf{I} = \begin{bmatrix} \alpha & 0 \\ 0 & \alpha \end{bmatrix}$
I.e, $w_i$ is drawn from a Gaussian density with variance $\alpha$ $$w_i \sim \mathcal{N}(0,\alpha)$$
interactive(children=(FloatSlider(value=0.5, description='alpha', max=1.0, min=0.1), Checkbox(value=False, des…
Sampling from the prior (weight space)¶
We can sample from the prior distribution to see what form we are imposing on the functions a priori (before seeing any data).
- Draw $w$ (left) independently from a Gaussian density $\mathbf{w} \sim \mathcal{N}(\mathbf{0}, \alpha\mathbf{I})$
- Use any normally distributed sampling technique, e.g. Box-Mueller transform
- Every sample yields a polynomial function $f(\mathbf{x})$ (right): $f(\mathbf{x}) = \mathbf{w} \boldsymbol{\phi}(\mathbf{x}).$
- For example, with $\boldsymbol{\phi}(\mathbf{x})$ being a polynomial:
interactive(children=(FloatSlider(value=2.1, description='alpha', max=5.0, min=0.1, step=0.5), IntSlider(value…
Learning Gaussian distributions¶
We assume that our data is Gaussian distributed: $P(y|x_{test},\textbf{X}) = \mathcal{N}(\mathbf{\mu,\Sigma})$
Example with learned mean $[m,m]$ and covariance $\begin{bmatrix} \alpha & \beta \\ \beta & \alpha \end{bmatrix}$
- The blue curve is the predicted $P(y|x_{test},\textbf{X})$
interactive(children=(FloatSlider(value=0.0, description='x_test', max=3.0, min=-3.0, step=0.5), FloatSlider(v…
Understanding covariances¶
- If two variables $x_i$ covariate strongly, knowing about $x_1$ tells us a lot about $x_2$
- If covariance is 0, knowing $x_1$ tells us nothing about $x_2$ (the conditional and marginal distributions are the same)
- For covariance matrix $\begin{bmatrix} 1 & \beta \\ \beta & 1 \end{bmatrix}$:
interactive(children=(FloatSlider(value=0.0, description='x1', max=3.0, min=-3.0, step=0.5), FloatSlider(value…
Sampling from higher-dimensional distributions¶
- Instead of sampling $\mathbf{w}$ and then multiplying by $\boldsymbol{\Phi}$, we can also generate examples of $f(x)$ directly.
- $\mathbf{f}$ with $n$ values can be sampled from an $n$-dimensional Gaussian distribution with zero mean and covariance matrix $\mathbf{K} = \alpha \boldsymbol{\Phi}\boldsymbol{\Phi}^\top$:
- $\mathbf{f}$ is a stochastic process: series of normally distributed variables (interpolated in the plot)
interactive(children=(FloatSlider(value=1.0, description='alpha', max=2.0, min=0.1), IntSlider(value=51, descr…
Repeat for 40 dimensions, with $\boldsymbol{\Phi}$ the polynomial transform:
Noisy functions¶
We normally add Gaussian noise to obtain our observations: $$ \mathbf{y} = \mathbf{f} + \boldsymbol{\epsilon} $$
interactive(children=(FloatSlider(value=0.05, description='sigma', max=0.1, min=0.01, step=0.01), Output()), _…
Gaussian Process¶
- Usually, we want our functions to be smooth: if two points are similar/nearby, the predictions should be similar.
- Hence, we need a similarity measure (a kernel)
- In a Gaussian process we can do this by specifying the covariance function directly (not as $\mathbf{K} = \alpha \boldsymbol{\Phi}\boldsymbol{\Phi}^\top$)
- The covariance matrix is simply the kernel matrix: $\mathbf{f} \sim \mathcal{N}(\mathbf{0},\mathbf{K})$
- The RBF (Gaussian) covariance function (or kernel) is specified by
where $\left\Vert\mathbf{x} - \mathbf{x}^\prime\right\Vert^2$ is the squared distance between the two input vectors
$$ \left\Vert\mathbf{x} - \mathbf{x}^\prime\right\Vert^2 = (\mathbf{x} - \mathbf{x}^\prime)^\top (\mathbf{x} - \mathbf{x}^\prime) $$and the length parameter $l$ controls the smoothness of the function and $\alpha$ the vertical variation.
Now the influence of a point decreases smoothly but exponentially
- These are our priors $P(y) = \mathcal{N}(\mathbf{0,\mathbf{K}})$, with mean 0
- We now want to condition it on our training data: $P(y|x_{test},\textbf{X}) = \mathcal{N}(\mathbf{\mu,\Sigma})$
interactive(children=(FloatSlider(value=1.0, description='alpha', max=2.0, min=0.1), IntSlider(value=10, descr…
Computing the posterior $P(\mathbf{y}|\mathbf{X})$¶
Assuming that $P(X)$ is a Gaussian density with a covariance given by kernel matrix $\mathbf{K}$, the model likelihood becomes: $$ P(\mathbf{y}|\mathbf{X}) = \frac{P(y) \ P(\mathbf{X} \mid y)}{P(\mathbf{X})} = \frac{1}{(2\pi)^{\frac{n}{2}}|\mathbf{K}|^{\frac{1}{2}}} \exp\left(-\frac{1}{2}\mathbf{y}^\top \left(\mathbf{K}+\sigma^2 \mathbf{I}\right)^{-1}\mathbf{y}\right) $$
Hence, the negative log likelihood (the objective function) is given by: $$ E(\boldsymbol{\theta}) = \frac{1}{2} \log |\mathbf{K}| + \frac{1}{2} \mathbf{y}^\top \left(\mathbf{K} + \sigma^2\mathbf{I}\right)^{-1}\mathbf{y} $$
The model parameters (e.g. noise variance $\sigma^2$) and the kernel parameters (e.g. lengthscale, variance) can be embedded in the covariance function and learned from data.
Good news: This loss function can be optimized using linear algebra (Cholesky Decomposition)
- Bad news: This is cubic in the number of data points AND the number of features: $\mathcal{O}(n^3 d^3)$
Making predictions¶
The model makes predictions for $\mathbf{f}$ that are unaffected by future values of $\mathbf{f}^*$.
If we think of $\mathbf{f}^*$ as test points, we can still write down a joint probability density over the training observations, $\mathbf{f}$ and the test observations, $\mathbf{f}^*$.
This joint probability density will be Gaussian, with a covariance matrix given by our kernel function, $k(\mathbf{x}_i, \mathbf{x}_j)$. $$ \begin{bmatrix}\mathbf{f} \\ \mathbf{f}^*\end{bmatrix} \sim \mathcal{N}\left(\mathbf{0}, \begin{bmatrix} \mathbf{K} & \mathbf{K}_\ast \\ \mathbf{K}_\ast^\top & \mathbf{K}_{\ast,\ast}\end{bmatrix}\right) $$
where $\mathbf{K}$ is the kernel matrix computed between all the training points,
$\mathbf{K}_\ast$ is the kernel matrix computed between the training points and the test points,
$\mathbf{K}_{\ast,\ast}$ is the kernel matrix computed between all the tests points and themselves.
Conditional Density $P(\mathbf{y}|x_{test} , \mathbf{X})$¶
Finally, we need to define conditional distributions to answer particular questions of interest
We will need the conditional density for making predictions. $$ \mathbf{f}^* | \mathbf{y} \sim \mathcal{N}(\boldsymbol{\mu}_f,\mathbf{C}_f) $$ with a mean given by $ \boldsymbol{\mu}_f = \mathbf{K}_*^\top \left[\mathbf{K} + \sigma^2 \mathbf{I}\right]^{-1} \mathbf{y} $
and a covariance given by $ \mathbf{C}_f = \mathbf{K}_{*,*} - \mathbf{K}_*^\top \left[\mathbf{K} + \sigma^2 \mathbf{I}\right]^{-1} \mathbf{K}_\ast. $
interactive(children=(IntSlider(value=13, description='nr_points', max=27), Output()), _dom_classes=('widget-i…
Remember that our prediction is the sum of the mean and the variance: $P(\mathbf{y}|x_{test} , \mathbf{X}) = \mathcal{N}(\mathbf{\mu,\Sigma})$
- The mean is the same as the one computed with kernel ridge (if given the same kernel and hyperparameters)
- The Gaussian process learned the covariance and the hyperparameters
interactive(children=(IntSlider(value=13, description='nr_points', max=27), Checkbox(value=False, description=…
The values on the diagonal of the covariance matrix give us the variance, so we can simply plot the mean and 95% confidence interval
interactive(children=(IntSlider(value=13, description='nr_points', max=27), IntSlider(value=12, description='l…
Gaussian Processes in practice (with GPy)¶
GPyRegressionGenerate a kernel first
- State the dimensionality of your input data
Variance and lengthscale are optional, default = 1
kernel = GPy.kern.RBF(input_dim=1, variance=1., lengthscale=1.)
Other kernels:
GPy.kern.BasisFuncKernel?
Build model:
m = GPy.models.GPRegression(X,Y,kernel)
Matern is a generalized RBF kernel that can scale between RBF and Exponential
Build the untrained GP. The shaded region corresponds to ~95% confidence intervals (i.e. +/- 2 standard deviation)
Train the model (optimize the parameters): maximize the likelihood of the data.
Best to optimize with a few restarts: the optimizer may converges to the high-noise solution. The optimizer is then restarted with a few random initialization of the parameter values.
You can also show results in 2D
We can plot 2D slices using the fixed_inputs argument to the plot function.
fixed_inputs is a list of tuples containing which of the inputs to fix, and to which value.
Gaussian Processes with scikit-learn¶
GaussianProcessRegressor- Hyperparameters:
kernel: kernel specifying the covariance function of the GP- Default: "1.0 * RBF(1.0)"
- Typically leave at default. Will be optimized during fitting
alpha: regularization parameter- Tikhonov regularization of covariance between the training points.
- Adds a (small) value to diagonal of the kernel matrix during fitting.
- Larger values:
- correspond to increased noise level in the observations
- also reduce potential numerical issues during fitting
- Default: 1e-10
n_restarts_optimizer: number of restarts of the optimizer- Default: 0. Best to do at least a few iterations.
- Optimizer finds kernel parameters maximizing log-marginal likelihood
- Retrieve predictions and confidence interval after fitting:
y_pred, sigma = gp.predict(x, return_std=True)
Example
Example with noisy data
Gaussian processes: Conclusions¶
Advantages:
- The prediction is probabilistic (Gaussian) so that one can compute empirical confidence intervals.
- The prediction interpolates the observations (at least for regular kernels).
- Versatile: different kernels can be specified.
Disadvantages:
- They are typically not sparse, i.e., they use the whole sample/feature information to perform the prediction.
- Sparse GPs also exist: they remember only the most important points
- They lose efficiency in high dimensional spaces – namely when the number of features exceeds a few dozens.
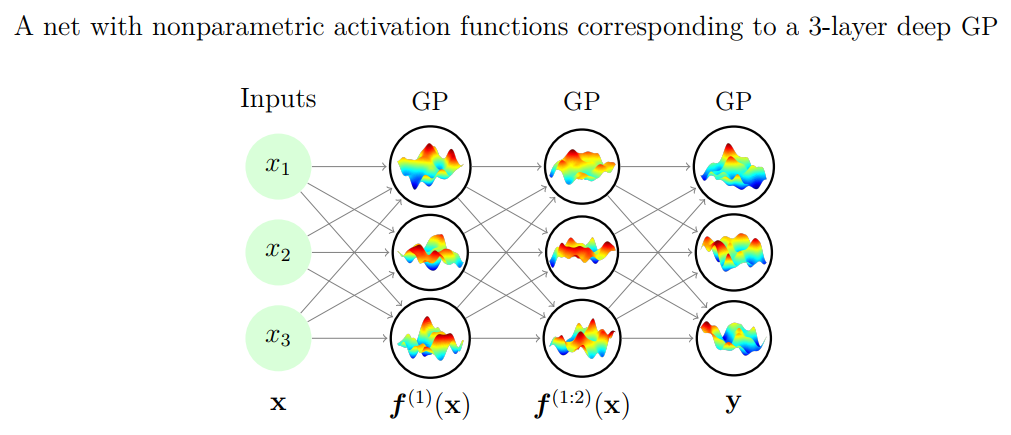
Gaussian processes and neural networks¶
- You can prove that a Gaussian process is equivalent to a neural network with one layer and an infinite number of nodes
- You can build deep Gaussian Processes by constructing layers of GPs
Bayesian optimization¶
- The incremental updates you can do with Bayesian models allow a more effective way to optimize functions
- E.g. to optimize the hyperparameter settings of a machine learning algorithm/pipeline
- After a number of random search iterations we know more about the performance of hyperparameter settings on the given dataset
- We can use this data to train a model, and predict which other hyperparameter values might be useful
- More generally, this is called model-based optimization
- This model is called a surrogate model
- This is often a probabilistic (e.g. Bayesian) model that predicts confidence intervals for all hyperparameter settings
- We use the predictions of this model to choose the next point to evaluate
- With every new evaluation, we update the surrogate model and repeat
Example (see figure):¶
- Consider only 1 continuous hyperparameter (X-axis)
- You can also do this for many more hyperparameters
- Y-axis shows cross-validation performance
- Evaluate a number of random hyperparameter settings (black dots)
- Sometimes an initialization design is used
- Train a model, and predict the expected performance of other (unseen) hyperparameter values
- Mean value (black line) and distribution (blue band)
- An acquisition function (green line) trades off maximal expected performace and maximal uncertainty
- Exploitation vs exploration
- Optimal value of the asquisition function is the next hyperparameter setting to be evaluated
- Repeat a fixed number of times, or until time budget runs out
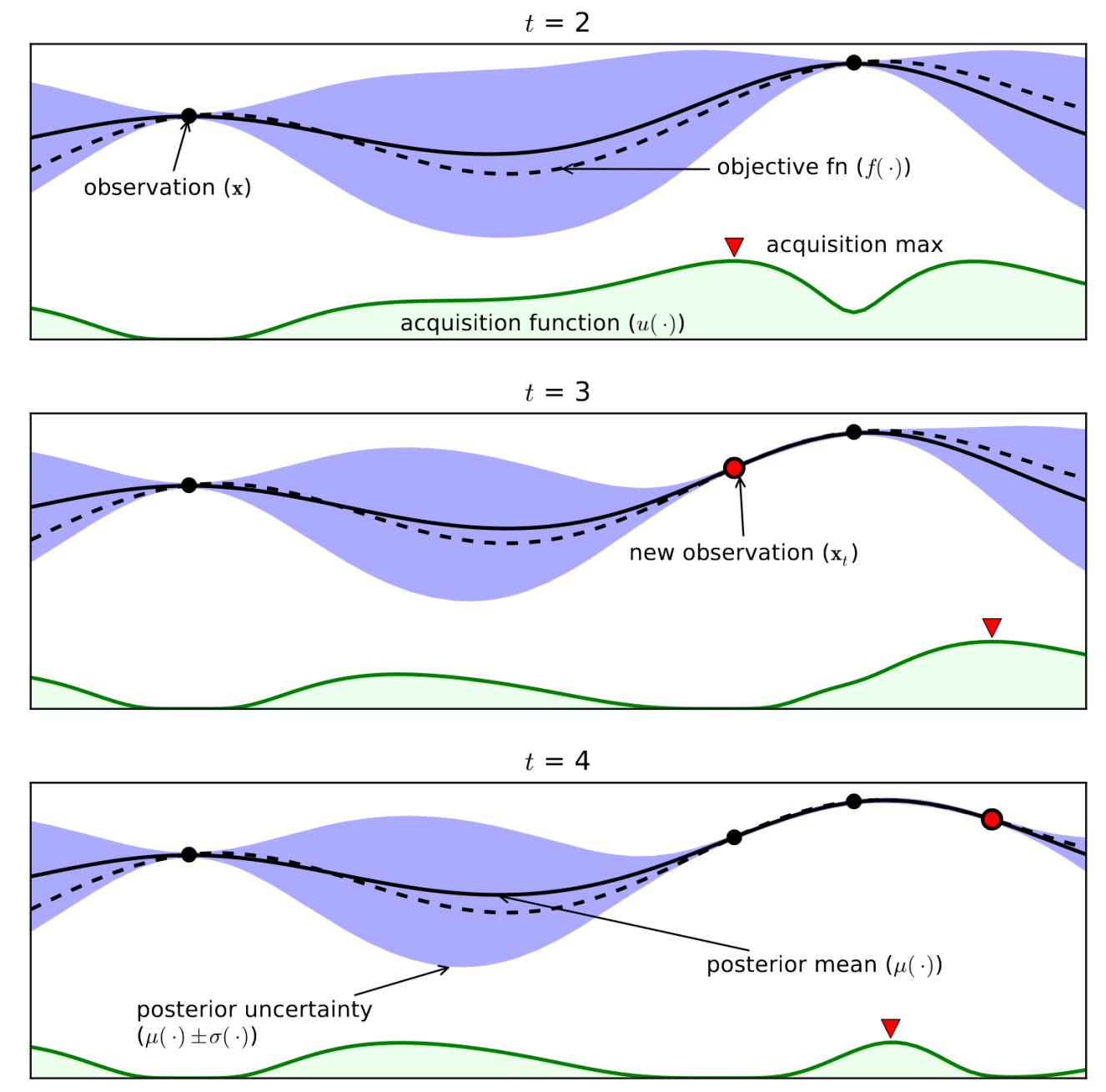
Shahriari et al. Taking the Human Out of the Loop: A Review of Bayesian Optimization
In 2 dimensions:
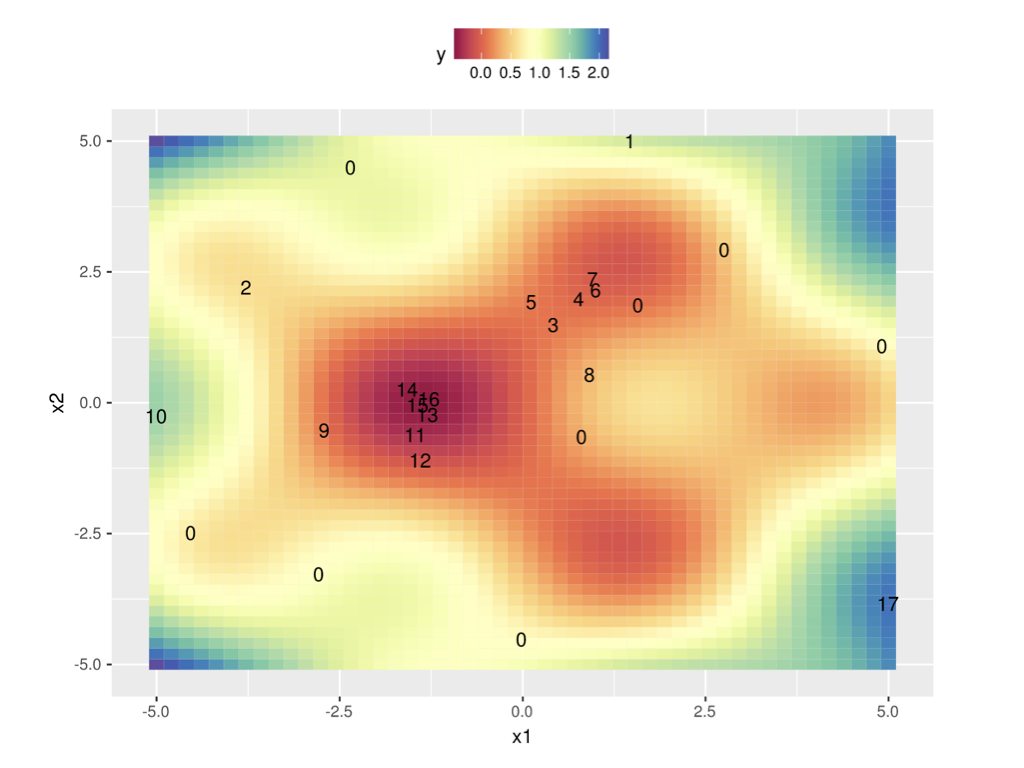
Surrogate models¶
- Surrogate model can be anything as long as it can do regression and is probabilistic
- Gaussian Processes are commonly used
- Smooth, good extrapolation, but don't scale well to many hyperparameters (cubic)
- Sparse GPs: select ‘inducing points’ that minimize info loss, more scalable
- Multi-task GPs: transfer surrogate models from other tasks
- Random Forests
- A lot more scalable, but don't extrapolate well
- Often an interpolation between predictions is used instead of the raw (step-wise) predictions
- Bayesian Neural Networks:
- Expensive, sensitive to hyperparameters
Acquisition Functions¶
- When we have trained the surrogate model, we ask it to predict a number of samples
- Can be simply random sampling
- Better: Thompson sampling
- fit a Gaussian distribution (a mixture of Gaussians) over the sampled points
- sample new points close to the means of the fitted Gaussians
- Typical acquisition function: Expected Improvement
- Models the predicted performance as a Gaussian distribution with the predicted mean and standard deviation
- Computes the expected performance improvement over the previous best configuration $\mathbf{X^+}$: $$EI(X) := \mathbb{E}\left[ \max\{0, f(\mathbf{X^+}) - f_{t+1}(\mathbf{X}) \} \right]$$
- Computing the expected performance requires an integration over the posterior distribution, but has a closed form solution.
Bayesian Optimization: conclusions¶
- More efficient way to optimize hyperparameters
- More similar to what humans would do
- Harder to parallellize
- Choice of surrogate model depends on your search space
- Very active research area
- For very high-dimensional search spaces, random forests are popular