Feature Maps¶
Linear models: $\hat{y} = \mathbf{w}\mathbf{x} + w_0 = \sum_{i=1}^{p} w_i x_i + w_0 = w_0 + w_1 x_1 + ... + w_p x_p $
When we cannot fit the data well, we can add non-linear transformations of the features
- Feature map (or basis expansion ) $\phi$ : $ X \rightarrow \mathbb{R}^d $
- E.g. Polynomial feature map: all polynomials up to degree $d$ and all products
- Example with $p=1, d=3$ :
Ridge regression example¶
Weights: [0.418]
- Add all polynomials $x^d$ up to degree 10 and fit again:
- e.g. use sklearn
PolynomialFeatures
- e.g. use sklearn
| x0 | x0^2 | x0^3 | x0^4 | x0^5 | x0^6 | x0^7 | x0^8 | x0^9 | x0^10 | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | -0.752759 | 0.566647 | -0.426548 | 0.321088 | -0.241702 | 0.181944 | -0.136960 | 0.103098 | -0.077608 | 0.058420 |
| 1 | 2.704286 | 7.313162 | 19.776880 | 53.482337 | 144.631526 | 391.124988 | 1057.713767 | 2860.360362 | 7735.232021 | 20918.278410 |
| 2 | 1.391964 | 1.937563 | 2.697017 | 3.754150 | 5.225640 | 7.273901 | 10.125005 | 14.093639 | 19.617834 | 27.307312 |
| 3 | 0.591951 | 0.350406 | 0.207423 | 0.122784 | 0.072682 | 0.043024 | 0.025468 | 0.015076 | 0.008924 | 0.005283 |
| 4 | -2.063888 | 4.259634 | -8.791409 | 18.144485 | -37.448187 | 77.288869 | -159.515582 | 329.222321 | -679.478050 | 1402.366700 |
Weights: [ 0.643 0.297 -0.69 -0.264 0.41 0.096 -0.076 -0.014 0.004 0.001]
How expensive is this?¶
- You may need MANY dimensions to fit the data
- Memory and computational cost
- More weights to learn, more likely overfitting
Ridge has a closed-form solution which we can compute with linear algebra: $$w^{*} = (X^{T}X + \alpha I)^{-1} X^T Y$$
Since X has $n$ rows (examples), and $d$ columns (features), $X^{T}X$ has dimensionality $d x d$
- Hence Ridge is quadratic in the number of features, $\mathcal{O}(d^2n)$
After the feature map $\Phi$, we get $$w^{*} = (\Phi(X)^{T}\Phi(X) + \alpha I)^{-1} \Phi(X)^T Y$$
Since $\Phi$ increases $d$ a lot, $\Phi(X)^{T}\Phi(X)$ becomes huge
Linear SVM example (classification)¶
We can add a new feature by taking the squares of feature1 values
Now we can fit a linear model
As a function of the original features, the decision boundary is now a polynomial as well $$y = w_0 + w_1 x_1 + w_2 x_2 + w_3 x_2^2 > 0$$
The kernel trick¶
- Computations in explicit, high-dimensional feature maps are expensive
- For some feature maps, we can, however, compute distances between points cheaply
- Without explicitly constructing the high-dimensional space at all
- Example: quadratic feature map for $\mathbf{x} = (x_1,..., x_p )$:
- A kernel function exists for this feature map to compute dot products
- Skip computation of $\Phi(x_i)$ and $\Phi(x_j)$ and compute $k(x_i,x_j)$ directly
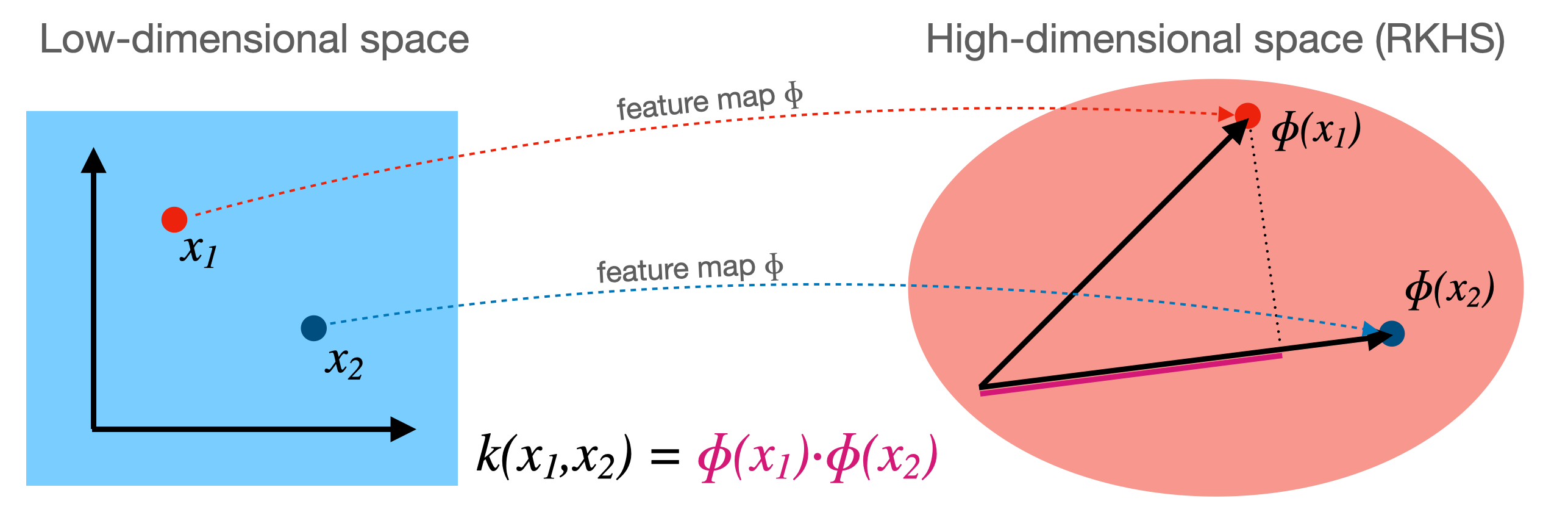
Kernelization¶
- Kernel $k$ corresponding to a feature map $\Phi$: $ k(\mathbf{x_i},\mathbf{x_j}) = \Phi(\mathbf{x_i}) \cdot \Phi(\mathbf{x_j})$
- Computes dot product between $x_i,x_j$ in a high-dimensional space $\mathcal{H}$
- Kernels are sometimes called generalized dot products
- $\mathcal{H}$ is called the reproducing kernel Hilbert space (RKHS)
- The dot product is a measure of the similarity between $x_i,x_j$
- Hence, a kernel can be seen as a similarity measure for high-dimensional spaces
- If we have a loss function based on dot products $\mathbf{x_i}\cdot\mathbf{x_j}$ it can be kernelized
- Simply replace the dot products with $k(\mathbf{x_i},\mathbf{x_j})$
Example: SVMs¶
- Linear SVMs (dual form, for $l$ support vectors with dual coefficients $a_i$ and classes $y_i$): $$\mathcal{L}_{Dual} (a_i) = \sum_{i=1}^{l} a_i - \frac{1}{2} \sum_{i,j=1}^{l} a_i a_j y_i y_j (\mathbf{x_i} . \mathbf{x_j}) $$
- Kernelized SVM, using any existing kernel $k$ we want: $$\mathcal{L}_{Dual} (a_i, k) = \sum_{i=1}^{l} a_i - \frac{1}{2} \sum_{i,j=1}^{l} a_i a_j y_i y_j k(\mathbf{x_i},\mathbf{x_j}) $$
1
Which kernels exist?¶
A (Mercer) kernel is any function $k: X \times X \rightarrow \mathbb{R}$ with these properties:
- Symmetry: $k(\mathbf{x_1},\mathbf{x_2}) = k(\mathbf{x_2},\mathbf{x_1}) \,\,\, \forall \mathbf{x_1},\mathbf{x_2} \in X$
- Positive definite: the kernel matrix $K$ is positive semi-definite
- Intuitively, $k(\mathbf{x_1},\mathbf{x_2}) \geq 0$
The kernel matrix (or Gram matrix) for $n$ points of $x_1,..., x_n \in X$ is defined as:
- Once computed ($\mathcal{O}(n^2)$), simply lookup $k(\mathbf{x_1}, \mathbf{x_2})$ for any two points
- In practice, you can either supply a kernel function or precompute the kernel matrix
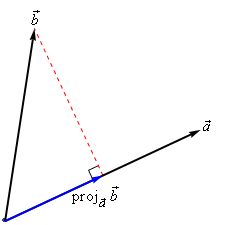
Linear kernel¶
- Input space is same as output space: $X = \mathcal{H} = \mathbb{R}^d$
- Feature map $\Phi(\mathbf{x}) = \mathbf{x}$
- Kernel: $ k_{linear}(\mathbf{x_i},\mathbf{x_j}) = \mathbf{x_i} \cdot \mathbf{x_j}$
- Geometrically, the dot product is the projection of $\mathbf{x_j}$ on hyperplane defined by $\mathbf{x_i}$
- Becomes larger if $\mathbf{x_i}$ and $\mathbf{x_j}$ are in the same 'direction'
- Linear kernel between point (0,1) and another unit vector an angle $a$ (in radians)
- Points with similar angles are deemed similar
Polynomial kernel¶
- If $k_1$, $k_2$ are kernels, then $\lambda . k_1$ ($\lambda \geq 0$), $k_1 + k_2$, and $k_1 . k_2$ are also kernels
- The polynomial kernel (for degree $d \in \mathbb{N}$) reproduces the polynomial feature map
- $\gamma$ is a scaling hyperparameter (default $\frac{1}{p}$)
- $c_0$ is a hyperparameter (default 1) to trade off influence of higher-order terms $$k_{poly}(\mathbf{x_1},\mathbf{x_2}) = (\gamma (\mathbf{x_1} \cdot \mathbf{x_2}) + c_0)^d$$
interactive(children=(IntSlider(value=5, description='degree', max=10, min=1), FloatSlider(value=1.0, descript…
RBF (Gaussian) kernel¶
The Radial Basis Function (RBF) feature map builds the Taylor series expansion of $e^x$ $$\Phi(x) = e^{-x^2/2\gamma^2} \Big[ 1, \sqrt{\frac{1}{1!\gamma^2}}x,\sqrt{\frac{1}{2!\gamma^4}}x^2,\sqrt{\frac{1}{3!\gamma^6}}x^3,\ldots\Big]^T$$
RBF (or Gaussian ) kernel with kernel width $\gamma \geq 0$:
$$k_{RBF}(\mathbf{x_1},\mathbf{x_2}) = exp(-\gamma ||\mathbf{x_1} - \mathbf{x_2}||^2)$$
interactive(children=(FloatSlider(value=4.91, description='gamma', max=10.0, min=0.01), Output()), _dom_classe…
- The RBF kernel $k_{RBF}(\mathbf{x_1},\mathbf{x_2}) = exp(-\gamma ||\mathbf{x_1} - \mathbf{x_2}||^2)$ does not use a dot product
- It only considers the distance between $\mathbf{x_1}$ and $\mathbf{x_2}$
- It's a local kernel : every data point only influences data points nearby
- linear and polynomial kernels are global : every point affects the whole space
- Similarity depends on closeness of points and kernel width
- value goes up for closer points and wider kernels (larger overlap)
interactive(children=(FloatSlider(value=0.5, description='gamma', max=1.0, min=0.1), FloatSlider(value=0.0, de…
Kernelized SVMs in practice¶
- You can use SVMs with any kernel to learn non-linear decision boundaries
SVM with RBF kernel¶
- Every support vector locally influences predictions, according to kernel width ($\gamma$)
- The prediction for test point $\mathbf{u}$: sum of the remaining influence of each support vector
- $f(x) = \sum_{i=1}^{l} a_i y_i k(\mathbf{x_i},\mathbf{u})$
interactive(children=(FloatSlider(value=4.6, description='gamma', max=10.0, min=0.1, step=0.5), FloatSlider(va…
Tuning RBF SVMs¶
- gamma (kernel width)
- high values cause narrow Gaussians, more support vectors, overfitting
- low values cause wide Gaussians, underfitting
- C (cost of margin violations)
- high values punish margin violations, cause narrow margins, overfitting
- low values cause wider margins, more support vectors, underfitting
Kernel overview
SVMs in practice¶
- C and gamma always need to be tuned
- Interacting regularizers. Find a good C, then finetune gamma
- SVMs expect all features to be approximately on the same scale
- Data needs to be scaled beforehand
- Allow to learn complex decision boundaries, even with few features
- Work well on both low- and high dimensional data
- Especially good at small, high-dimensional data
- Hard to inspect, although support vectors can be inspected
- In sklearn, you can use
SVCfor classification with a range of kernelsSVRfor regression
Other kernels¶
- There are many more possible kernels
- If no kernel function exists, we can still precompute the kernel matrix
- All you need is some similarity measure, and you can use SVMs
- Text kernels:
- Word kernels: build a bag-of-words representation of the text (e.g. TFIDF)
- Kernel is the inner product between these vectors
- Subsequence kernels: sequences are similar if they share many sub-sequences
- Build a kernel matrix based on pairwise similarities
- Word kernels: build a bag-of-words representation of the text (e.g. TFIDF)
- Graph kernels: Same idea (e.g. find common subgraphs to measure similarity)
- These days, deep learning embeddings are more frequently used
The Representer Theorem¶
- We can kernelize many other loss functions as well
The Representer Theorem states that if we have a loss function $\mathcal{L}'$ with
- $\mathcal{L}$ an arbitrary loss function using some function $f$ of the inputs $\mathbf{x}$
- $\mathcal{R}$ a (non-decreasing) regularization score (e.g. L1 or L2) and constant $\lambda$ $$\mathcal{\mathcal{L}'}(\mathbf{w}) = \mathcal{L}(y,f(\mathbf{x})) + \lambda \mathcal{R} (||\mathbf{w}||)$$
Then the weights $\mathbf{w}$ can be described as a linear combination of the training samples: $$\mathbf{w} = \sum_{i=1}^{n} a_i y_i f(\mathbf{x_i})$$
- Note that this is exactly what we found for SVMs: $ \mathbf{w} = \sum_{i=1}^{l} a_i y_i \mathbf{\mathbf{x_i}} $
- Hence, we can also kernelize Ridge regression, Logistic regression, Perceptrons, Support Vector Regression, ...
Kernelized Ridge regression¶
- The linear Ridge regression loss (with $\mathbf{x_0}=1$): $$\mathcal{L}_{Ridge}(\mathbf{w}) = \sum_{i=0}^{n} (y_i-\mathbf{w}\mathbf{x_i})^2 + \lambda \| w \|^2$$
- Filling in $\mathbf{w} = \sum_{i=1}^{n} \alpha_i y_i \mathbf{x_i}$ yields the dual formulation: $$\mathcal{L}_{Ridge}(\mathbf{w}) = \sum_{i=1}^{n} (y_i-\sum_{j=1}^{n} \alpha_j y_j \mathbf{x_i}\mathbf{x_j})^2 + \lambda \sum_{i=1}^{n} \sum_{j=1}^{n} \alpha_i \alpha_j y_i y_j \mathbf{x_i}\mathbf{x_j}$$
- Generalize $\mathbf{x_i}\cdot\mathbf{x_j}$ to $k(\mathbf{x_i},\mathbf{x_j})$ $$\mathcal{L}_{KernelRidge}(\mathbf{\alpha},k) = \sum_{i=1}^{n} (y_i-\sum_{j=1}^{n} \alpha_j y_j k(\mathbf{x_i},\mathbf{x_n}))^2 + \lambda \sum_{i=1}^{n} \sum_{j=1}^{n} \alpha_i \alpha_j y_i y_j k(\mathbf{x_i},\mathbf{x_j})$$
Example of kernelized Ridge¶
- Prediction (red) is now a linear combination of kernels (blue): $y = \sum_{j=1}^{n} \alpha_j y_j k(\mathbf{x},\mathbf{x_j})$
- We learn a dual coefficient for each point
interactive(children=(FloatSlider(value=1.0, description='gamma', max=2.0, min=0.1), FloatSlider(value=0.0, de…
- Fitting our regression data with
KernelRidge
interactive(children=(FloatSlider(value=4.51, description='gamma', max=10.0, min=0.01, step=0.5), Output()), _…
Other kernelized methods¶
- Same procedure can be done for logistic regression
- For perceptrons, $\alpha \rightarrow \alpha+1$ after every misclassification $$\mathcal{L}_{DualPerceptron}(x_i,k) = max(0,y_i \sum_{j=1}^{n} \alpha_j y_j k(\mathbf{x_j},\mathbf{x_i}))$$
- Support Vector Regression behaves similarly to Kernel Ridge
interactive(children=(FloatSlider(value=4.51, description='gamma', max=10.0, min=0.01, step=0.5), Output()), _…
Summary¶
- Feature maps $\Phi(x)$ transform features to create a higher-dimensional space
- Allows learning non-linear functions or boundaries, but very expensive/slow
- For some $\Phi(x)$, we can compute dot products without constructing this space
- Kernel trick: $k(\mathbf{x_i},\mathbf{x_j}) = \Phi(\mathbf{x_i}) \cdot \Phi(\mathbf{x_j})$
- Kernel $k$ (generalized dot product) is a measure of similarity between $\mathbf{x_i}$ and $\mathbf{x_j}$
- There are many such kernels
- Polynomial kernel: $k_{poly}(\mathbf{x_1},\mathbf{x_2}) = (\gamma (\mathbf{x_1} \cdot \mathbf{x_2}) + c_0)^d$
- RBF (Gaussian) kernel: $k_{RBF}(\mathbf{x_1},\mathbf{x_2}) = exp(-\gamma ||\mathbf{x_1} - \mathbf{x_2}||^2)$
- A kernel matrix can be precomputed using any similarity measure (e.g. for text, graphs,...)
- Any loss function where inputs appear only as dot products can be kernelized
- E.g. Linear SVMs: simply replace the dot product with a kernel of choice
- The Representer theorem states which other loss functions can also be kernelized and how
- Ridge regression, Logistic regression, Perceptrons,...